
Papaya meleira virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 8.43
Get precalculated fractions of proteins
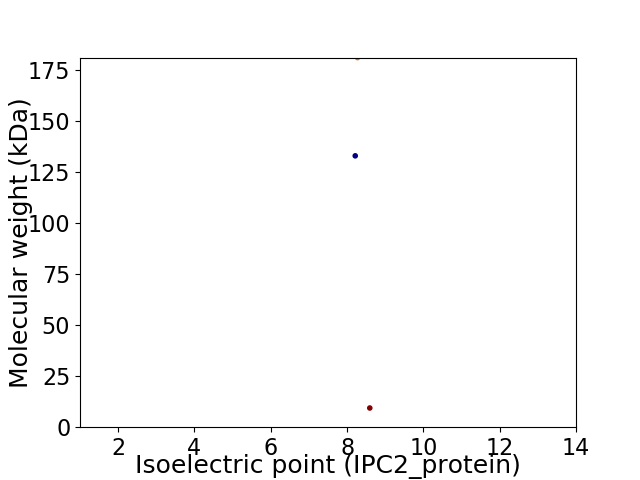
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0QL14|A0A0P0QL14_9VIRU RNA-directed RNA polymerase OS=Papaya meleira virus OX=1497848 GN=pol PE=3 SV=1
MM1 pKa = 7.5AASQGRR7 pKa = 11.84YY8 pKa = 9.13CEE10 pKa = 3.99EE11 pKa = 3.63RR12 pKa = 11.84FNLFKK17 pKa = 10.66EE18 pKa = 4.32YY19 pKa = 10.51TSLNEE24 pKa = 4.83FPDD27 pKa = 3.81LNLKK31 pKa = 10.14VEE33 pKa = 4.26EE34 pKa = 4.25TFKK37 pKa = 11.44NFVFKK42 pKa = 10.9LSEE45 pKa = 4.13IEE47 pKa = 4.2GLPLLSHH54 pKa = 7.0CNLLIDD60 pKa = 4.87IKK62 pKa = 11.19GFLDD66 pKa = 3.87KK67 pKa = 10.96FPLIHH72 pKa = 6.88YY73 pKa = 8.03DD74 pKa = 3.6YY75 pKa = 11.2NNSNFRR81 pKa = 11.84NIVTWIFIEE90 pKa = 3.97PLPRR94 pKa = 11.84EE95 pKa = 4.18IYY97 pKa = 10.27SYY99 pKa = 11.55LPVKK103 pKa = 10.39LFNTINIFILKK114 pKa = 9.84DD115 pKa = 3.57LEE117 pKa = 4.6KK118 pKa = 10.23EE119 pKa = 3.94LPKK122 pKa = 10.57INFISNLILNKK133 pKa = 10.4NNLEE137 pKa = 3.78ITNNLFFKK145 pKa = 10.73PLGRR149 pKa = 11.84LGGKK153 pKa = 6.24THH155 pKa = 7.39LLIGDD160 pKa = 4.21FLKK163 pKa = 11.05LFNFSFSDD171 pKa = 3.12IGRR174 pKa = 11.84EE175 pKa = 3.52EE176 pKa = 4.49DD177 pKa = 3.07KK178 pKa = 11.15GKK180 pKa = 10.72KK181 pKa = 8.79VVHH184 pKa = 6.02ILINKK189 pKa = 8.38NRR191 pKa = 11.84VNNNIGIYY199 pKa = 10.45DD200 pKa = 4.07FNNLIKK206 pKa = 10.74LNKK209 pKa = 9.03DD210 pKa = 3.53TILSNDD216 pKa = 3.51IIYY219 pKa = 9.22TLISVLLYY227 pKa = 10.26QLEE230 pKa = 4.48LGCEE234 pKa = 4.13EE235 pKa = 4.35SWNSLNVAGLLISPITSPLGFICTLFFISSNITKK269 pKa = 8.93LTILEE274 pKa = 3.99ASKK277 pKa = 10.34ICKK280 pKa = 9.85KK281 pKa = 10.16FQNTVRR287 pKa = 11.84IGGRR291 pKa = 11.84LPIFNIPITDD301 pKa = 3.67DD302 pKa = 3.07NKK304 pKa = 10.87YY305 pKa = 10.7FLDD308 pKa = 4.17GQKK311 pKa = 10.86GILSIEE317 pKa = 4.86GIRR320 pKa = 11.84MLYY323 pKa = 10.63GIDD326 pKa = 3.64SLLGPSEE333 pKa = 4.4FYY335 pKa = 10.67KK336 pKa = 10.65YY337 pKa = 10.9DD338 pKa = 4.9PISEE342 pKa = 4.36IITRR346 pKa = 11.84QVHH349 pKa = 6.1PNLEE353 pKa = 3.84LSLYY357 pKa = 10.5KK358 pKa = 10.7GGSRR362 pKa = 11.84SNKK365 pKa = 9.39AYY367 pKa = 10.49SSTIEE372 pKa = 3.9AVNKK376 pKa = 9.7RR377 pKa = 11.84EE378 pKa = 3.94EE379 pKa = 4.38IIRR382 pKa = 11.84QDD384 pKa = 3.26MYY386 pKa = 11.29NSLFFSVVKK395 pKa = 10.47KK396 pKa = 8.9LHH398 pKa = 6.52RR399 pKa = 11.84PFKK402 pKa = 10.25TFRR405 pKa = 11.84EE406 pKa = 4.09WFNIKK411 pKa = 10.21FSWSASGGCPGAKK424 pKa = 9.85VFWSEE429 pKa = 4.1SEE431 pKa = 4.03KK432 pKa = 10.68IRR434 pKa = 11.84VNKK437 pKa = 10.19RR438 pKa = 11.84GALLNVKK445 pKa = 9.83VDD447 pKa = 4.15DD448 pKa = 3.95VVKK451 pKa = 10.83AINEE455 pKa = 4.41TYY457 pKa = 10.56NNPVHH462 pKa = 6.08YY463 pKa = 10.45SKK465 pKa = 11.04AARR468 pKa = 11.84KK469 pKa = 8.91YY470 pKa = 8.46EE471 pKa = 3.9KK472 pKa = 10.67GKK474 pKa = 10.12NRR476 pKa = 11.84VIWNTTLLMYY486 pKa = 10.3LAQSYY491 pKa = 10.61LLYY494 pKa = 9.98TFEE497 pKa = 6.42SILDD501 pKa = 3.93PLQHH505 pKa = 6.92DD506 pKa = 4.68FSFEE510 pKa = 4.08LKK512 pKa = 10.5QISSWNSSTANADD525 pKa = 2.91MRR527 pKa = 11.84FISNQEE533 pKa = 3.75RR534 pKa = 11.84LRR536 pKa = 11.84SLKK539 pKa = 10.8DD540 pKa = 3.19NSGLMFDD547 pKa = 4.57YY548 pKa = 11.23SDD550 pKa = 4.25FNINHH555 pKa = 6.69SIVVQNKK562 pKa = 9.75LYY564 pKa = 10.86DD565 pKa = 3.56VLSQVLFNKK574 pKa = 9.69MKK576 pKa = 10.6VNQEE580 pKa = 3.72NYY582 pKa = 9.79FQVLKK587 pKa = 10.31DD588 pKa = 3.5IKK590 pKa = 10.39WITEE594 pKa = 3.93YY595 pKa = 10.63ILRR598 pKa = 11.84AKK600 pKa = 8.99TFSVLDD606 pKa = 3.73AGVTDD611 pKa = 4.26NYY613 pKa = 10.2VSQVMRR619 pKa = 11.84SLEE622 pKa = 4.0SGEE625 pKa = 4.49RR626 pKa = 11.84GTSFINTFLSNCYY639 pKa = 9.5IRR641 pKa = 11.84NLNTNSRR648 pKa = 11.84SLFGRR653 pKa = 11.84QLLINNLWLQGDD665 pKa = 4.34DD666 pKa = 3.49VFAMTKK672 pKa = 10.1NLEE675 pKa = 4.57DD676 pKa = 3.49GCLAVNLFNCLGYY689 pKa = 10.63AGQPSKK695 pKa = 10.84IGLSLYY701 pKa = 11.07NNGEE705 pKa = 4.31FLRR708 pKa = 11.84LSYY711 pKa = 10.91DD712 pKa = 3.48PSCEE716 pKa = 3.97IIGGYY721 pKa = 8.69PIRR724 pKa = 11.84STSGLISGEE733 pKa = 3.94FFKK736 pKa = 11.06EE737 pKa = 4.1SIYY740 pKa = 11.28DD741 pKa = 3.59PDD743 pKa = 3.79TRR745 pKa = 11.84AGCYY749 pKa = 8.85LSSVIRR755 pKa = 11.84CNVRR759 pKa = 11.84GAIIPNNLIYY769 pKa = 10.28ILCQRR774 pKa = 11.84NCRR777 pKa = 11.84LIYY780 pKa = 8.85TKK782 pKa = 10.42PNSKK786 pKa = 10.3EE787 pKa = 4.01KK788 pKa = 10.36IVMIGDD794 pKa = 3.65PVLALIPSILGGFGINSILDD814 pKa = 3.2IGMKK818 pKa = 9.84HH819 pKa = 5.17QVPGTDD825 pKa = 2.82QLLNRR830 pKa = 11.84LQLSPKK836 pKa = 8.72MISGGWFSSKK846 pKa = 9.87FKK848 pKa = 10.83NLNPPKK854 pKa = 10.17DD855 pKa = 3.7SLPIGEE861 pKa = 5.07LFWIRR866 pKa = 11.84RR867 pKa = 11.84VSNNSNNNFNSYY879 pKa = 10.31YY880 pKa = 10.6VINTTKK886 pKa = 10.56SNPILRR892 pKa = 11.84FDD894 pKa = 5.2NIPDD898 pKa = 3.4ILGDD902 pKa = 3.5KK903 pKa = 10.27SIPDD907 pKa = 3.69FLSAYY912 pKa = 6.97TICNKK917 pKa = 10.35SLTKK921 pKa = 10.47EE922 pKa = 3.47ITQNIVKK929 pKa = 9.87SSVMGALPLDD939 pKa = 4.38LLSKK943 pKa = 10.76SLTKK947 pKa = 10.79YY948 pKa = 11.0GEE950 pKa = 4.19DD951 pKa = 3.24QLEE954 pKa = 4.03YY955 pKa = 10.17RR956 pKa = 11.84SKK958 pKa = 11.54CNVHH962 pKa = 4.24WVKK965 pKa = 9.72YY966 pKa = 7.49TSNFGSLNIKK976 pKa = 10.32LSFISHH982 pKa = 6.69YY983 pKa = 9.76ISNFIKK989 pKa = 10.49FLISDD994 pKa = 4.65EE995 pKa = 4.46IKK997 pKa = 9.49TSHH1000 pKa = 7.15KK1001 pKa = 10.17IDD1003 pKa = 2.97ISVNLNYY1010 pKa = 10.89GILPTLFIEE1019 pKa = 4.83YY1020 pKa = 10.32GFSLQEE1026 pKa = 3.71NFEE1029 pKa = 4.19RR1030 pKa = 11.84VIDD1033 pKa = 3.99EE1034 pKa = 4.89LNTKK1038 pKa = 9.68EE1039 pKa = 4.25LLEE1042 pKa = 5.1LIASKK1047 pKa = 7.55QTKK1050 pKa = 9.48KK1051 pKa = 10.86FSITSFFKK1059 pKa = 11.05NILILNKK1066 pKa = 10.29YY1067 pKa = 6.55NTKK1070 pKa = 10.68LYY1072 pKa = 10.63LKK1074 pKa = 8.9WIKK1077 pKa = 10.67GKK1079 pKa = 10.03ILLFPPVFSLIDD1091 pKa = 3.5SDD1093 pKa = 5.78IINIWRR1099 pKa = 11.84GWSLFSVEE1107 pKa = 5.05KK1108 pKa = 10.35MILHH1112 pKa = 6.1GVNLRR1117 pKa = 11.84EE1118 pKa = 4.5DD1119 pKa = 4.99DD1120 pKa = 3.36IPYY1123 pKa = 10.16IILEE1127 pKa = 4.27LEE1129 pKa = 4.16LLIEE1133 pKa = 4.29NYY1135 pKa = 9.98IKK1137 pKa = 10.82DD1138 pKa = 3.73SLEE1141 pKa = 3.91SLNKK1145 pKa = 9.36INKK1148 pKa = 8.62FYY1150 pKa = 11.56SMQTKK1155 pKa = 10.07AA1156 pKa = 4.14
MM1 pKa = 7.5AASQGRR7 pKa = 11.84YY8 pKa = 9.13CEE10 pKa = 3.99EE11 pKa = 3.63RR12 pKa = 11.84FNLFKK17 pKa = 10.66EE18 pKa = 4.32YY19 pKa = 10.51TSLNEE24 pKa = 4.83FPDD27 pKa = 3.81LNLKK31 pKa = 10.14VEE33 pKa = 4.26EE34 pKa = 4.25TFKK37 pKa = 11.44NFVFKK42 pKa = 10.9LSEE45 pKa = 4.13IEE47 pKa = 4.2GLPLLSHH54 pKa = 7.0CNLLIDD60 pKa = 4.87IKK62 pKa = 11.19GFLDD66 pKa = 3.87KK67 pKa = 10.96FPLIHH72 pKa = 6.88YY73 pKa = 8.03DD74 pKa = 3.6YY75 pKa = 11.2NNSNFRR81 pKa = 11.84NIVTWIFIEE90 pKa = 3.97PLPRR94 pKa = 11.84EE95 pKa = 4.18IYY97 pKa = 10.27SYY99 pKa = 11.55LPVKK103 pKa = 10.39LFNTINIFILKK114 pKa = 9.84DD115 pKa = 3.57LEE117 pKa = 4.6KK118 pKa = 10.23EE119 pKa = 3.94LPKK122 pKa = 10.57INFISNLILNKK133 pKa = 10.4NNLEE137 pKa = 3.78ITNNLFFKK145 pKa = 10.73PLGRR149 pKa = 11.84LGGKK153 pKa = 6.24THH155 pKa = 7.39LLIGDD160 pKa = 4.21FLKK163 pKa = 11.05LFNFSFSDD171 pKa = 3.12IGRR174 pKa = 11.84EE175 pKa = 3.52EE176 pKa = 4.49DD177 pKa = 3.07KK178 pKa = 11.15GKK180 pKa = 10.72KK181 pKa = 8.79VVHH184 pKa = 6.02ILINKK189 pKa = 8.38NRR191 pKa = 11.84VNNNIGIYY199 pKa = 10.45DD200 pKa = 4.07FNNLIKK206 pKa = 10.74LNKK209 pKa = 9.03DD210 pKa = 3.53TILSNDD216 pKa = 3.51IIYY219 pKa = 9.22TLISVLLYY227 pKa = 10.26QLEE230 pKa = 4.48LGCEE234 pKa = 4.13EE235 pKa = 4.35SWNSLNVAGLLISPITSPLGFICTLFFISSNITKK269 pKa = 8.93LTILEE274 pKa = 3.99ASKK277 pKa = 10.34ICKK280 pKa = 9.85KK281 pKa = 10.16FQNTVRR287 pKa = 11.84IGGRR291 pKa = 11.84LPIFNIPITDD301 pKa = 3.67DD302 pKa = 3.07NKK304 pKa = 10.87YY305 pKa = 10.7FLDD308 pKa = 4.17GQKK311 pKa = 10.86GILSIEE317 pKa = 4.86GIRR320 pKa = 11.84MLYY323 pKa = 10.63GIDD326 pKa = 3.64SLLGPSEE333 pKa = 4.4FYY335 pKa = 10.67KK336 pKa = 10.65YY337 pKa = 10.9DD338 pKa = 4.9PISEE342 pKa = 4.36IITRR346 pKa = 11.84QVHH349 pKa = 6.1PNLEE353 pKa = 3.84LSLYY357 pKa = 10.5KK358 pKa = 10.7GGSRR362 pKa = 11.84SNKK365 pKa = 9.39AYY367 pKa = 10.49SSTIEE372 pKa = 3.9AVNKK376 pKa = 9.7RR377 pKa = 11.84EE378 pKa = 3.94EE379 pKa = 4.38IIRR382 pKa = 11.84QDD384 pKa = 3.26MYY386 pKa = 11.29NSLFFSVVKK395 pKa = 10.47KK396 pKa = 8.9LHH398 pKa = 6.52RR399 pKa = 11.84PFKK402 pKa = 10.25TFRR405 pKa = 11.84EE406 pKa = 4.09WFNIKK411 pKa = 10.21FSWSASGGCPGAKK424 pKa = 9.85VFWSEE429 pKa = 4.1SEE431 pKa = 4.03KK432 pKa = 10.68IRR434 pKa = 11.84VNKK437 pKa = 10.19RR438 pKa = 11.84GALLNVKK445 pKa = 9.83VDD447 pKa = 4.15DD448 pKa = 3.95VVKK451 pKa = 10.83AINEE455 pKa = 4.41TYY457 pKa = 10.56NNPVHH462 pKa = 6.08YY463 pKa = 10.45SKK465 pKa = 11.04AARR468 pKa = 11.84KK469 pKa = 8.91YY470 pKa = 8.46EE471 pKa = 3.9KK472 pKa = 10.67GKK474 pKa = 10.12NRR476 pKa = 11.84VIWNTTLLMYY486 pKa = 10.3LAQSYY491 pKa = 10.61LLYY494 pKa = 9.98TFEE497 pKa = 6.42SILDD501 pKa = 3.93PLQHH505 pKa = 6.92DD506 pKa = 4.68FSFEE510 pKa = 4.08LKK512 pKa = 10.5QISSWNSSTANADD525 pKa = 2.91MRR527 pKa = 11.84FISNQEE533 pKa = 3.75RR534 pKa = 11.84LRR536 pKa = 11.84SLKK539 pKa = 10.8DD540 pKa = 3.19NSGLMFDD547 pKa = 4.57YY548 pKa = 11.23SDD550 pKa = 4.25FNINHH555 pKa = 6.69SIVVQNKK562 pKa = 9.75LYY564 pKa = 10.86DD565 pKa = 3.56VLSQVLFNKK574 pKa = 9.69MKK576 pKa = 10.6VNQEE580 pKa = 3.72NYY582 pKa = 9.79FQVLKK587 pKa = 10.31DD588 pKa = 3.5IKK590 pKa = 10.39WITEE594 pKa = 3.93YY595 pKa = 10.63ILRR598 pKa = 11.84AKK600 pKa = 8.99TFSVLDD606 pKa = 3.73AGVTDD611 pKa = 4.26NYY613 pKa = 10.2VSQVMRR619 pKa = 11.84SLEE622 pKa = 4.0SGEE625 pKa = 4.49RR626 pKa = 11.84GTSFINTFLSNCYY639 pKa = 9.5IRR641 pKa = 11.84NLNTNSRR648 pKa = 11.84SLFGRR653 pKa = 11.84QLLINNLWLQGDD665 pKa = 4.34DD666 pKa = 3.49VFAMTKK672 pKa = 10.1NLEE675 pKa = 4.57DD676 pKa = 3.49GCLAVNLFNCLGYY689 pKa = 10.63AGQPSKK695 pKa = 10.84IGLSLYY701 pKa = 11.07NNGEE705 pKa = 4.31FLRR708 pKa = 11.84LSYY711 pKa = 10.91DD712 pKa = 3.48PSCEE716 pKa = 3.97IIGGYY721 pKa = 8.69PIRR724 pKa = 11.84STSGLISGEE733 pKa = 3.94FFKK736 pKa = 11.06EE737 pKa = 4.1SIYY740 pKa = 11.28DD741 pKa = 3.59PDD743 pKa = 3.79TRR745 pKa = 11.84AGCYY749 pKa = 8.85LSSVIRR755 pKa = 11.84CNVRR759 pKa = 11.84GAIIPNNLIYY769 pKa = 10.28ILCQRR774 pKa = 11.84NCRR777 pKa = 11.84LIYY780 pKa = 8.85TKK782 pKa = 10.42PNSKK786 pKa = 10.3EE787 pKa = 4.01KK788 pKa = 10.36IVMIGDD794 pKa = 3.65PVLALIPSILGGFGINSILDD814 pKa = 3.2IGMKK818 pKa = 9.84HH819 pKa = 5.17QVPGTDD825 pKa = 2.82QLLNRR830 pKa = 11.84LQLSPKK836 pKa = 8.72MISGGWFSSKK846 pKa = 9.87FKK848 pKa = 10.83NLNPPKK854 pKa = 10.17DD855 pKa = 3.7SLPIGEE861 pKa = 5.07LFWIRR866 pKa = 11.84RR867 pKa = 11.84VSNNSNNNFNSYY879 pKa = 10.31YY880 pKa = 10.6VINTTKK886 pKa = 10.56SNPILRR892 pKa = 11.84FDD894 pKa = 5.2NIPDD898 pKa = 3.4ILGDD902 pKa = 3.5KK903 pKa = 10.27SIPDD907 pKa = 3.69FLSAYY912 pKa = 6.97TICNKK917 pKa = 10.35SLTKK921 pKa = 10.47EE922 pKa = 3.47ITQNIVKK929 pKa = 9.87SSVMGALPLDD939 pKa = 4.38LLSKK943 pKa = 10.76SLTKK947 pKa = 10.79YY948 pKa = 11.0GEE950 pKa = 4.19DD951 pKa = 3.24QLEE954 pKa = 4.03YY955 pKa = 10.17RR956 pKa = 11.84SKK958 pKa = 11.54CNVHH962 pKa = 4.24WVKK965 pKa = 9.72YY966 pKa = 7.49TSNFGSLNIKK976 pKa = 10.32LSFISHH982 pKa = 6.69YY983 pKa = 9.76ISNFIKK989 pKa = 10.49FLISDD994 pKa = 4.65EE995 pKa = 4.46IKK997 pKa = 9.49TSHH1000 pKa = 7.15KK1001 pKa = 10.17IDD1003 pKa = 2.97ISVNLNYY1010 pKa = 10.89GILPTLFIEE1019 pKa = 4.83YY1020 pKa = 10.32GFSLQEE1026 pKa = 3.71NFEE1029 pKa = 4.19RR1030 pKa = 11.84VIDD1033 pKa = 3.99EE1034 pKa = 4.89LNTKK1038 pKa = 9.68EE1039 pKa = 4.25LLEE1042 pKa = 5.1LIASKK1047 pKa = 7.55QTKK1050 pKa = 9.48KK1051 pKa = 10.86FSITSFFKK1059 pKa = 11.05NILILNKK1066 pKa = 10.29YY1067 pKa = 6.55NTKK1070 pKa = 10.68LYY1072 pKa = 10.63LKK1074 pKa = 8.9WIKK1077 pKa = 10.67GKK1079 pKa = 10.03ILLFPPVFSLIDD1091 pKa = 3.5SDD1093 pKa = 5.78IINIWRR1099 pKa = 11.84GWSLFSVEE1107 pKa = 5.05KK1108 pKa = 10.35MILHH1112 pKa = 6.1GVNLRR1117 pKa = 11.84EE1118 pKa = 4.5DD1119 pKa = 4.99DD1120 pKa = 3.36IPYY1123 pKa = 10.16IILEE1127 pKa = 4.27LEE1129 pKa = 4.16LLIEE1133 pKa = 4.29NYY1135 pKa = 9.98IKK1137 pKa = 10.82DD1138 pKa = 3.73SLEE1141 pKa = 3.91SLNKK1145 pKa = 9.36INKK1148 pKa = 8.62FYY1150 pKa = 11.56SMQTKK1155 pKa = 10.07AA1156 pKa = 4.14
Molecular weight: 132.88 kDa
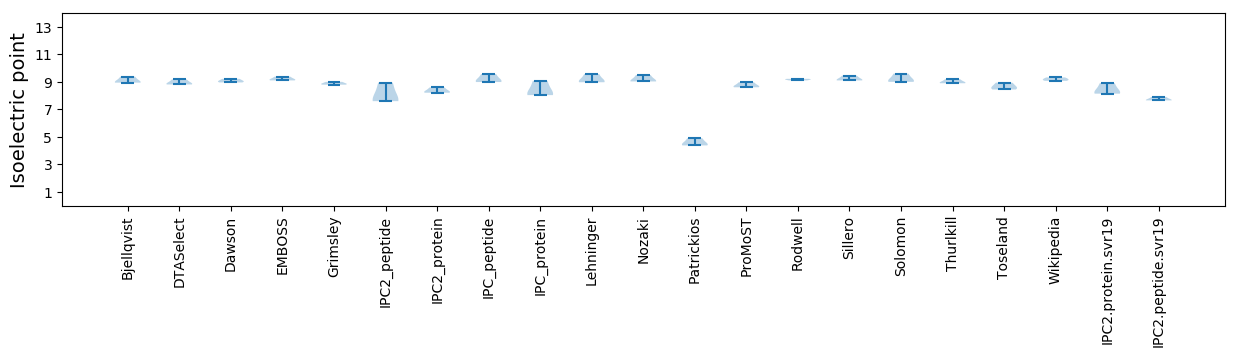
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0QKV0|A0A0P0QKV0_9VIRU Putative structural protein OS=Papaya meleira virus OX=1497848 GN=gag PE=4 SV=1
MM1 pKa = 7.41VEE3 pKa = 4.51RR4 pKa = 11.84LTGMTNGFCHH14 pKa = 6.89RR15 pKa = 11.84LNSPALNRR23 pKa = 11.84GRR25 pKa = 11.84PILLSNCYY33 pKa = 10.09LPLSNFSSNRR43 pKa = 11.84SKK45 pKa = 11.13DD46 pKa = 2.81IGRR49 pKa = 11.84ATRR52 pKa = 11.84LAFEE56 pKa = 5.12LPDD59 pKa = 3.86LLSPSFLISLTLHH72 pKa = 6.37FEE74 pKa = 3.67AHH76 pKa = 6.94FISFVYY82 pKa = 10.15II83 pKa = 3.73
MM1 pKa = 7.41VEE3 pKa = 4.51RR4 pKa = 11.84LTGMTNGFCHH14 pKa = 6.89RR15 pKa = 11.84LNSPALNRR23 pKa = 11.84GRR25 pKa = 11.84PILLSNCYY33 pKa = 10.09LPLSNFSSNRR43 pKa = 11.84SKK45 pKa = 11.13DD46 pKa = 2.81IGRR49 pKa = 11.84ATRR52 pKa = 11.84LAFEE56 pKa = 5.12LPDD59 pKa = 3.86LLSPSFLISLTLHH72 pKa = 6.37FEE74 pKa = 3.67AHH76 pKa = 6.94FISFVYY82 pKa = 10.15II83 pKa = 3.73
Molecular weight: 9.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2835 |
83 |
1596 |
945.0 |
107.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.633 ± 0.8 | 1.199 ± 0.317 |
4.656 ± 0.541 | 5.15 ± 0.368 |
5.115 ± 1.073 | 5.15 ± 0.144 |
2.046 ± 0.597 | 8.536 ± 1.207 |
6.772 ± 1.414 | 10.265 ± 2.09 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.623 ± 0.258 | 8.289 ± 0.517 |
5.256 ± 1.069 | 3.069 ± 0.882 |
4.656 ± 0.997 | 8.924 ± 0.759 |
4.974 ± 0.544 | 5.397 ± 0.994 |
1.199 ± 0.287 | 4.092 ± 0.414 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
