
Acer yangbiense
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Sapindaceae; Hippocastanoideae; Acereae; Acer
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
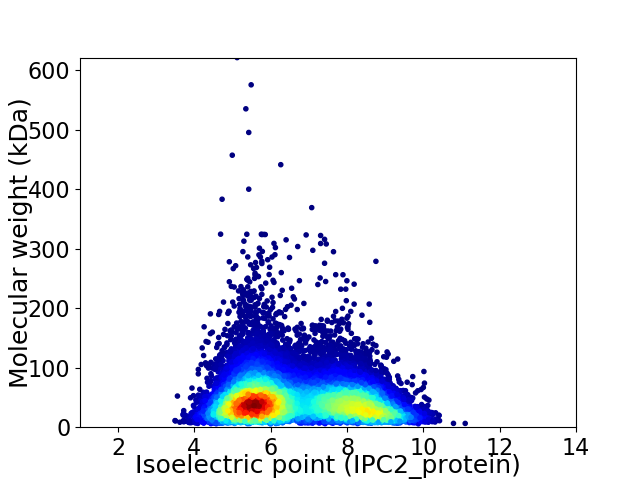
Virtual 2D-PAGE plot for 28112 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C7I7V0|A0A5C7I7V0_9ROSI Uncharacterized protein OS=Acer yangbiense OX=1000413 GN=EZV62_006928 PE=4 SV=1
MM1 pKa = 7.47YY2 pKa = 7.74EE3 pKa = 4.11TNHH6 pKa = 7.35LEE8 pKa = 4.13EE9 pKa = 5.15NPPSAYY15 pKa = 10.46GQMDD19 pKa = 4.04SCPGNVCPDD28 pKa = 3.55FDD30 pKa = 4.72GNQVFHH36 pKa = 7.44SPIPEE41 pKa = 4.48CGASFPNLEE50 pKa = 4.22YY51 pKa = 10.78SSPLPEE57 pKa = 3.75MPIWRR62 pKa = 11.84TVEE65 pKa = 5.15DD66 pKa = 4.22IPSPSIPVDD75 pKa = 3.61DD76 pKa = 4.07SHH78 pKa = 9.03RR79 pKa = 11.84EE80 pKa = 3.93EE81 pKa = 5.25DD82 pKa = 4.13LHH84 pKa = 7.62TGDD87 pKa = 5.88AFALTDD93 pKa = 5.02DD94 pKa = 4.94GDD96 pKa = 4.36AKK98 pKa = 10.58DD99 pKa = 3.73TTGGAEE105 pKa = 4.42GYY107 pKa = 9.82LAEE110 pKa = 5.29LSNSLLNFNEE120 pKa = 4.29EE121 pKa = 4.1EE122 pKa = 5.08LMTLDD127 pKa = 4.59DD128 pKa = 4.4VEE130 pKa = 5.98KK131 pKa = 11.14EE132 pKa = 4.19MIDD135 pKa = 3.23KK136 pKa = 11.22SFFDD140 pKa = 4.58GLNLLLNSPNDD151 pKa = 3.45VNQDD155 pKa = 3.39HH156 pKa = 6.51MPSPEE161 pKa = 4.35PEE163 pKa = 3.99TSVTPDD169 pKa = 3.42YY170 pKa = 10.84LANAYY175 pKa = 8.81GSCPVEE181 pKa = 4.23SVEE184 pKa = 4.07NVHH187 pKa = 5.79MQSSASASDD196 pKa = 3.44PQFPEE201 pKa = 5.32LIDD204 pKa = 3.64GVIYY208 pKa = 8.88CTLNMEE214 pKa = 5.53DD215 pKa = 5.25PDD217 pKa = 4.3IPCNDD222 pKa = 3.84DD223 pKa = 3.55VFFPNKK229 pKa = 9.7LQPLSVSAVAQRR241 pKa = 11.84EE242 pKa = 4.47AGNSTCSSVKK252 pKa = 10.49DD253 pKa = 3.33LSGNKK258 pKa = 9.46NLTDD262 pKa = 4.14GGPVLLQRR270 pKa = 11.84EE271 pKa = 4.48QEE273 pKa = 4.26NPAQSQVSSQMMGSQVIPEE292 pKa = 4.3KK293 pKa = 10.97VQFHH297 pKa = 5.7PVKK300 pKa = 10.63FGLPNCDD307 pKa = 3.58SPHH310 pKa = 5.79LASRR314 pKa = 11.84KK315 pKa = 9.39PGIACGGSNLTNSMSICTDD334 pKa = 2.47SHH336 pKa = 6.14QPARR340 pKa = 11.84LKK342 pKa = 10.81EE343 pKa = 4.06EE344 pKa = 4.05NKK346 pKa = 9.82EE347 pKa = 3.74YY348 pKa = 10.49CYY350 pKa = 10.83KK351 pKa = 10.8RR352 pKa = 11.84NANGVKK358 pKa = 9.59QEE360 pKa = 4.44CDD362 pKa = 3.47DD363 pKa = 4.2PATIQDD369 pKa = 4.15CQTLNAEE376 pKa = 4.55FGSTNIPDD384 pKa = 4.65LEE386 pKa = 5.25PDD388 pKa = 3.14INDD391 pKa = 4.94PISEE395 pKa = 4.23PEE397 pKa = 4.0EE398 pKa = 3.85PSIEE402 pKa = 4.18SDD404 pKa = 3.88DD405 pKa = 3.99DD406 pKa = 3.61VPYY409 pKa = 11.11YY410 pKa = 11.05SDD412 pKa = 3.72IEE414 pKa = 4.52AMILDD419 pKa = 4.49MDD421 pKa = 5.92LDD423 pKa = 4.92PDD425 pKa = 3.87DD426 pKa = 4.11QDD428 pKa = 3.6IYY430 pKa = 9.78EE431 pKa = 4.36QEE433 pKa = 3.93VSKK436 pKa = 10.78YY437 pKa = 6.6QHH439 pKa = 6.45EE440 pKa = 4.16DD441 pKa = 2.92TRR443 pKa = 11.84RR444 pKa = 11.84GNHH447 pKa = 4.9QAA449 pKa = 3.52
MM1 pKa = 7.47YY2 pKa = 7.74EE3 pKa = 4.11TNHH6 pKa = 7.35LEE8 pKa = 4.13EE9 pKa = 5.15NPPSAYY15 pKa = 10.46GQMDD19 pKa = 4.04SCPGNVCPDD28 pKa = 3.55FDD30 pKa = 4.72GNQVFHH36 pKa = 7.44SPIPEE41 pKa = 4.48CGASFPNLEE50 pKa = 4.22YY51 pKa = 10.78SSPLPEE57 pKa = 3.75MPIWRR62 pKa = 11.84TVEE65 pKa = 5.15DD66 pKa = 4.22IPSPSIPVDD75 pKa = 3.61DD76 pKa = 4.07SHH78 pKa = 9.03RR79 pKa = 11.84EE80 pKa = 3.93EE81 pKa = 5.25DD82 pKa = 4.13LHH84 pKa = 7.62TGDD87 pKa = 5.88AFALTDD93 pKa = 5.02DD94 pKa = 4.94GDD96 pKa = 4.36AKK98 pKa = 10.58DD99 pKa = 3.73TTGGAEE105 pKa = 4.42GYY107 pKa = 9.82LAEE110 pKa = 5.29LSNSLLNFNEE120 pKa = 4.29EE121 pKa = 4.1EE122 pKa = 5.08LMTLDD127 pKa = 4.59DD128 pKa = 4.4VEE130 pKa = 5.98KK131 pKa = 11.14EE132 pKa = 4.19MIDD135 pKa = 3.23KK136 pKa = 11.22SFFDD140 pKa = 4.58GLNLLLNSPNDD151 pKa = 3.45VNQDD155 pKa = 3.39HH156 pKa = 6.51MPSPEE161 pKa = 4.35PEE163 pKa = 3.99TSVTPDD169 pKa = 3.42YY170 pKa = 10.84LANAYY175 pKa = 8.81GSCPVEE181 pKa = 4.23SVEE184 pKa = 4.07NVHH187 pKa = 5.79MQSSASASDD196 pKa = 3.44PQFPEE201 pKa = 5.32LIDD204 pKa = 3.64GVIYY208 pKa = 8.88CTLNMEE214 pKa = 5.53DD215 pKa = 5.25PDD217 pKa = 4.3IPCNDD222 pKa = 3.84DD223 pKa = 3.55VFFPNKK229 pKa = 9.7LQPLSVSAVAQRR241 pKa = 11.84EE242 pKa = 4.47AGNSTCSSVKK252 pKa = 10.49DD253 pKa = 3.33LSGNKK258 pKa = 9.46NLTDD262 pKa = 4.14GGPVLLQRR270 pKa = 11.84EE271 pKa = 4.48QEE273 pKa = 4.26NPAQSQVSSQMMGSQVIPEE292 pKa = 4.3KK293 pKa = 10.97VQFHH297 pKa = 5.7PVKK300 pKa = 10.63FGLPNCDD307 pKa = 3.58SPHH310 pKa = 5.79LASRR314 pKa = 11.84KK315 pKa = 9.39PGIACGGSNLTNSMSICTDD334 pKa = 2.47SHH336 pKa = 6.14QPARR340 pKa = 11.84LKK342 pKa = 10.81EE343 pKa = 4.06EE344 pKa = 4.05NKK346 pKa = 9.82EE347 pKa = 3.74YY348 pKa = 10.49CYY350 pKa = 10.83KK351 pKa = 10.8RR352 pKa = 11.84NANGVKK358 pKa = 9.59QEE360 pKa = 4.44CDD362 pKa = 3.47DD363 pKa = 4.2PATIQDD369 pKa = 4.15CQTLNAEE376 pKa = 4.55FGSTNIPDD384 pKa = 4.65LEE386 pKa = 5.25PDD388 pKa = 3.14INDD391 pKa = 4.94PISEE395 pKa = 4.23PEE397 pKa = 4.0EE398 pKa = 3.85PSIEE402 pKa = 4.18SDD404 pKa = 3.88DD405 pKa = 3.99DD406 pKa = 3.61VPYY409 pKa = 11.11YY410 pKa = 11.05SDD412 pKa = 3.72IEE414 pKa = 4.52AMILDD419 pKa = 4.49MDD421 pKa = 5.92LDD423 pKa = 4.92PDD425 pKa = 3.87DD426 pKa = 4.11QDD428 pKa = 3.6IYY430 pKa = 9.78EE431 pKa = 4.36QEE433 pKa = 3.93VSKK436 pKa = 10.78YY437 pKa = 6.6QHH439 pKa = 6.45EE440 pKa = 4.16DD441 pKa = 2.92TRR443 pKa = 11.84RR444 pKa = 11.84GNHH447 pKa = 4.9QAA449 pKa = 3.52
Molecular weight: 49.41 kDa
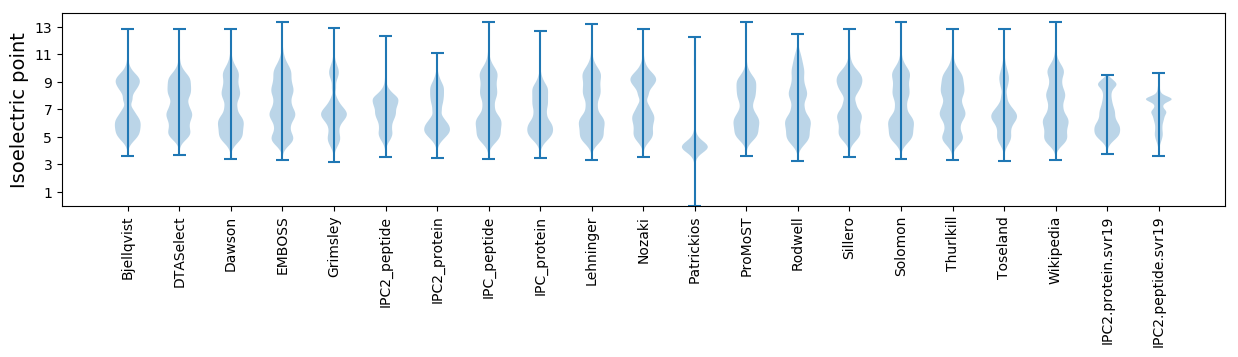
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C7GZY6|A0A5C7GZY6_9ROSI Uncharacterized protein OS=Acer yangbiense OX=1000413 GN=EZV62_022882 PE=4 SV=1
MM1 pKa = 7.71PFIKK5 pKa = 9.49FTKK8 pKa = 9.7NPSSSSINSTLLISFPLLHH27 pKa = 6.53SRR29 pKa = 11.84RR30 pKa = 11.84PSHH33 pKa = 6.7LLLRR37 pKa = 11.84STHH40 pKa = 5.05GRR42 pKa = 11.84RR43 pKa = 11.84PSHH46 pKa = 6.9RR47 pKa = 11.84IKK49 pKa = 10.33IRR51 pKa = 11.84NRR53 pKa = 11.84GVV55 pKa = 2.72
MM1 pKa = 7.71PFIKK5 pKa = 9.49FTKK8 pKa = 9.7NPSSSSINSTLLISFPLLHH27 pKa = 6.53SRR29 pKa = 11.84RR30 pKa = 11.84PSHH33 pKa = 6.7LLLRR37 pKa = 11.84STHH40 pKa = 5.05GRR42 pKa = 11.84RR43 pKa = 11.84PSHH46 pKa = 6.9RR47 pKa = 11.84IKK49 pKa = 10.33IRR51 pKa = 11.84NRR53 pKa = 11.84GVV55 pKa = 2.72
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12256278 |
51 |
5462 |
436.0 |
48.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.287 ± 0.014 | 1.948 ± 0.007 |
5.362 ± 0.009 | 6.438 ± 0.016 |
4.355 ± 0.01 | 6.374 ± 0.015 |
2.365 ± 0.006 | 5.584 ± 0.011 |
6.294 ± 0.013 | 9.815 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.502 ± 0.006 | 4.737 ± 0.01 |
4.637 ± 0.015 | 3.633 ± 0.011 |
5.052 ± 0.011 | 8.886 ± 0.015 |
4.93 ± 0.009 | 6.636 ± 0.009 |
1.325 ± 0.005 | 2.841 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
