
Aphanothece hegewaldii CCALA 016
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Aphanothecaceae; Aphanothece; Aphanothece hegewaldii
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
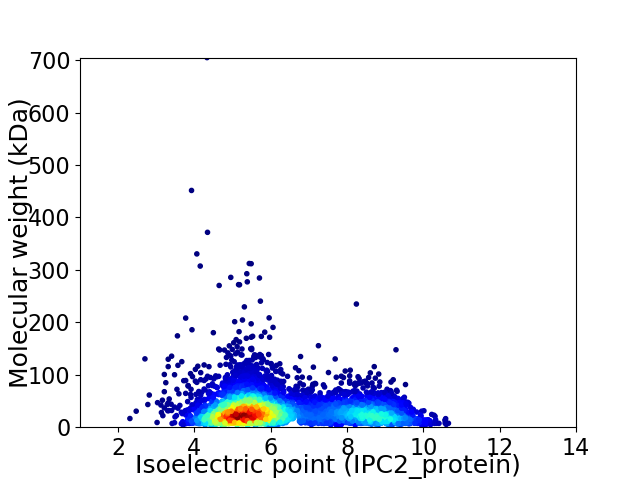
Virtual 2D-PAGE plot for 4827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1LX15|A0A2T1LX15_9CHRO Transposase OS=Aphanothece hegewaldii CCALA 016 OX=2107694 GN=C7H19_13395 PE=3 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84RR3 pKa = 11.84IGTIGPDD10 pKa = 3.26NLLGTSEE17 pKa = 4.96DD18 pKa = 5.04DD19 pKa = 3.17ILKK22 pKa = 10.25GLKK25 pKa = 10.67GNDD28 pKa = 3.18VLRR31 pKa = 11.84GLKK34 pKa = 10.4GDD36 pKa = 4.26DD37 pKa = 3.98LLFGGDD43 pKa = 3.91GNDD46 pKa = 3.42TIYY49 pKa = 11.16AGGGNDD55 pKa = 3.46KK56 pKa = 11.21VIGGEE61 pKa = 4.32GNDD64 pKa = 3.72FLYY67 pKa = 10.73KK68 pKa = 10.07ILSGKK73 pKa = 5.61TTAVNFSYY81 pKa = 10.92TNPEE85 pKa = 3.55VGKK88 pKa = 9.57IQGIEE93 pKa = 4.02SVYY96 pKa = 10.87LSTGLGNDD104 pKa = 4.23TIISTAAANTIYY116 pKa = 11.18SNDD119 pKa = 3.38GNDD122 pKa = 3.85WIMTGAGRR130 pKa = 11.84DD131 pKa = 4.0SINSGAGDD139 pKa = 4.23DD140 pKa = 4.64YY141 pKa = 11.19ISSGAGDD148 pKa = 3.95DD149 pKa = 4.77QIDD152 pKa = 3.57AGKK155 pKa = 11.22GNDD158 pKa = 3.66TVIGGAGNDD167 pKa = 3.47SLYY170 pKa = 11.04KK171 pKa = 10.65DD172 pKa = 5.13FSDD175 pKa = 3.24QTTGINFSYY184 pKa = 10.67TNPEE188 pKa = 3.6VGNIQGIEE196 pKa = 4.03AVTLSTGSGDD206 pKa = 3.55DD207 pKa = 4.06TIVLTAAEE215 pKa = 5.23GYY217 pKa = 9.93LYY219 pKa = 10.12HH220 pKa = 7.32KK221 pKa = 10.73NYY223 pKa = 10.08IYY225 pKa = 11.27SNDD228 pKa = 3.5GNNSITTGAGNEE240 pKa = 4.09QIISGAGDD248 pKa = 3.94DD249 pKa = 4.76YY250 pKa = 11.26ISSGAGDD257 pKa = 4.79DD258 pKa = 4.41YY259 pKa = 11.37ISSGTGNDD267 pKa = 3.49TVIGGAGNDD276 pKa = 3.56HH277 pKa = 7.27LYY279 pKa = 11.09KK280 pKa = 10.79DD281 pKa = 5.12FSDD284 pKa = 3.31QTTGINFSYY293 pKa = 10.65TDD295 pKa = 3.93PNVGNIQGIEE305 pKa = 4.1SLHH308 pKa = 6.57LSTGSGDD315 pKa = 3.59DD316 pKa = 4.24TIVSTAAVSLFSVYY330 pKa = 11.02SNYY333 pKa = 9.7ISSNGGNDD341 pKa = 3.96SITTGAGDD349 pKa = 4.35DD350 pKa = 3.92IIYY353 pKa = 10.62AGTGDD358 pKa = 3.6DD359 pKa = 3.7TVIGGEE365 pKa = 4.2GNDD368 pKa = 3.57YY369 pKa = 10.69LGKK372 pKa = 10.35SFSDD376 pKa = 3.19LTTAVNFSYY385 pKa = 10.88TDD387 pKa = 3.73PNSGKK392 pKa = 9.21IQGIEE397 pKa = 3.77RR398 pKa = 11.84LYY400 pKa = 11.1LSTGSGDD407 pKa = 3.56DD408 pKa = 4.24TIVSTAAVGFSDD420 pKa = 3.94YY421 pKa = 10.84SANEE425 pKa = 3.82IYY427 pKa = 11.02SNGGNDD433 pKa = 5.22SITTSAGNDD442 pKa = 3.83YY443 pKa = 10.89INSGAGNDD451 pKa = 4.85GIDD454 pKa = 4.13AGTGDD459 pKa = 3.83DD460 pKa = 3.82TVIGGEE466 pKa = 4.22GNDD469 pKa = 3.74YY470 pKa = 9.32LTKK473 pKa = 10.76DD474 pKa = 3.78FSNLTRR480 pKa = 11.84AVNFSYY486 pKa = 10.89TDD488 pKa = 3.73PNSGKK493 pKa = 9.21IQGIEE498 pKa = 3.77RR499 pKa = 11.84LYY501 pKa = 11.03LSTGAGDD508 pKa = 3.8DD509 pKa = 4.4TIVSAAVGFSNYY521 pKa = 9.28SANEE525 pKa = 3.73IYY527 pKa = 10.91SNEE530 pKa = 3.99GNDD533 pKa = 5.14SITTSAGDD541 pKa = 3.79DD542 pKa = 4.26FISSGAGNDD551 pKa = 4.06SITTGAGDD559 pKa = 3.94DD560 pKa = 4.87DD561 pKa = 4.74IYY563 pKa = 11.6SGAGNDD569 pKa = 4.7LIKK572 pKa = 10.17TGAGNDD578 pKa = 4.3GIIAGEE584 pKa = 4.32GNDD587 pKa = 3.86TLIGGLGNDD596 pKa = 3.81KK597 pKa = 9.93LTGNSGQDD605 pKa = 2.65QFVFNTPTEE614 pKa = 4.63GIDD617 pKa = 3.44RR618 pKa = 11.84IYY620 pKa = 11.04DD621 pKa = 3.7FSLIDD626 pKa = 3.94DD627 pKa = 5.31KK628 pKa = 11.45IAVSRR633 pKa = 11.84SGFSNDD639 pKa = 4.31LIAGAAITVAQFTIDD654 pKa = 3.55SVATTASQRR663 pKa = 11.84FIYY666 pKa = 10.48NSASGALFFDD676 pKa = 4.33ADD678 pKa = 4.35GVGSTAAIQFATLDD692 pKa = 3.54SGLALTNADD701 pKa = 3.29IFVTPP706 pKa = 4.59
MM1 pKa = 7.49RR2 pKa = 11.84RR3 pKa = 11.84IGTIGPDD10 pKa = 3.26NLLGTSEE17 pKa = 4.96DD18 pKa = 5.04DD19 pKa = 3.17ILKK22 pKa = 10.25GLKK25 pKa = 10.67GNDD28 pKa = 3.18VLRR31 pKa = 11.84GLKK34 pKa = 10.4GDD36 pKa = 4.26DD37 pKa = 3.98LLFGGDD43 pKa = 3.91GNDD46 pKa = 3.42TIYY49 pKa = 11.16AGGGNDD55 pKa = 3.46KK56 pKa = 11.21VIGGEE61 pKa = 4.32GNDD64 pKa = 3.72FLYY67 pKa = 10.73KK68 pKa = 10.07ILSGKK73 pKa = 5.61TTAVNFSYY81 pKa = 10.92TNPEE85 pKa = 3.55VGKK88 pKa = 9.57IQGIEE93 pKa = 4.02SVYY96 pKa = 10.87LSTGLGNDD104 pKa = 4.23TIISTAAANTIYY116 pKa = 11.18SNDD119 pKa = 3.38GNDD122 pKa = 3.85WIMTGAGRR130 pKa = 11.84DD131 pKa = 4.0SINSGAGDD139 pKa = 4.23DD140 pKa = 4.64YY141 pKa = 11.19ISSGAGDD148 pKa = 3.95DD149 pKa = 4.77QIDD152 pKa = 3.57AGKK155 pKa = 11.22GNDD158 pKa = 3.66TVIGGAGNDD167 pKa = 3.47SLYY170 pKa = 11.04KK171 pKa = 10.65DD172 pKa = 5.13FSDD175 pKa = 3.24QTTGINFSYY184 pKa = 10.67TNPEE188 pKa = 3.6VGNIQGIEE196 pKa = 4.03AVTLSTGSGDD206 pKa = 3.55DD207 pKa = 4.06TIVLTAAEE215 pKa = 5.23GYY217 pKa = 9.93LYY219 pKa = 10.12HH220 pKa = 7.32KK221 pKa = 10.73NYY223 pKa = 10.08IYY225 pKa = 11.27SNDD228 pKa = 3.5GNNSITTGAGNEE240 pKa = 4.09QIISGAGDD248 pKa = 3.94DD249 pKa = 4.76YY250 pKa = 11.26ISSGAGDD257 pKa = 4.79DD258 pKa = 4.41YY259 pKa = 11.37ISSGTGNDD267 pKa = 3.49TVIGGAGNDD276 pKa = 3.56HH277 pKa = 7.27LYY279 pKa = 11.09KK280 pKa = 10.79DD281 pKa = 5.12FSDD284 pKa = 3.31QTTGINFSYY293 pKa = 10.65TDD295 pKa = 3.93PNVGNIQGIEE305 pKa = 4.1SLHH308 pKa = 6.57LSTGSGDD315 pKa = 3.59DD316 pKa = 4.24TIVSTAAVSLFSVYY330 pKa = 11.02SNYY333 pKa = 9.7ISSNGGNDD341 pKa = 3.96SITTGAGDD349 pKa = 4.35DD350 pKa = 3.92IIYY353 pKa = 10.62AGTGDD358 pKa = 3.6DD359 pKa = 3.7TVIGGEE365 pKa = 4.2GNDD368 pKa = 3.57YY369 pKa = 10.69LGKK372 pKa = 10.35SFSDD376 pKa = 3.19LTTAVNFSYY385 pKa = 10.88TDD387 pKa = 3.73PNSGKK392 pKa = 9.21IQGIEE397 pKa = 3.77RR398 pKa = 11.84LYY400 pKa = 11.1LSTGSGDD407 pKa = 3.56DD408 pKa = 4.24TIVSTAAVGFSDD420 pKa = 3.94YY421 pKa = 10.84SANEE425 pKa = 3.82IYY427 pKa = 11.02SNGGNDD433 pKa = 5.22SITTSAGNDD442 pKa = 3.83YY443 pKa = 10.89INSGAGNDD451 pKa = 4.85GIDD454 pKa = 4.13AGTGDD459 pKa = 3.83DD460 pKa = 3.82TVIGGEE466 pKa = 4.22GNDD469 pKa = 3.74YY470 pKa = 9.32LTKK473 pKa = 10.76DD474 pKa = 3.78FSNLTRR480 pKa = 11.84AVNFSYY486 pKa = 10.89TDD488 pKa = 3.73PNSGKK493 pKa = 9.21IQGIEE498 pKa = 3.77RR499 pKa = 11.84LYY501 pKa = 11.03LSTGAGDD508 pKa = 3.8DD509 pKa = 4.4TIVSAAVGFSNYY521 pKa = 9.28SANEE525 pKa = 3.73IYY527 pKa = 10.91SNEE530 pKa = 3.99GNDD533 pKa = 5.14SITTSAGDD541 pKa = 3.79DD542 pKa = 4.26FISSGAGNDD551 pKa = 4.06SITTGAGDD559 pKa = 3.94DD560 pKa = 4.87DD561 pKa = 4.74IYY563 pKa = 11.6SGAGNDD569 pKa = 4.7LIKK572 pKa = 10.17TGAGNDD578 pKa = 4.3GIIAGEE584 pKa = 4.32GNDD587 pKa = 3.86TLIGGLGNDD596 pKa = 3.81KK597 pKa = 9.93LTGNSGQDD605 pKa = 2.65QFVFNTPTEE614 pKa = 4.63GIDD617 pKa = 3.44RR618 pKa = 11.84IYY620 pKa = 11.04DD621 pKa = 3.7FSLIDD626 pKa = 3.94DD627 pKa = 5.31KK628 pKa = 11.45IAVSRR633 pKa = 11.84SGFSNDD639 pKa = 4.31LIAGAAITVAQFTIDD654 pKa = 3.55SVATTASQRR663 pKa = 11.84FIYY666 pKa = 10.48NSASGALFFDD676 pKa = 4.33ADD678 pKa = 4.35GVGSTAAIQFATLDD692 pKa = 3.54SGLALTNADD701 pKa = 3.29IFVTPP706 pKa = 4.59
Molecular weight: 72.23 kDa
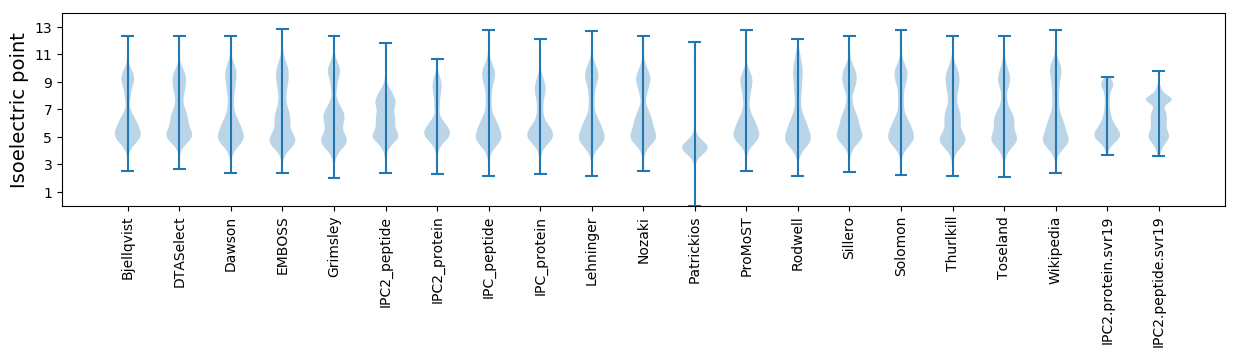
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T1M2L3|A0A2T1M2L3_9CHRO Haloacid dehalogenase OS=Aphanothece hegewaldii CCALA 016 OX=2107694 GN=C7H19_02655 PE=4 SV=1
MM1 pKa = 8.03PINIRR6 pKa = 11.84RR7 pKa = 11.84CISCHH12 pKa = 6.11KK13 pKa = 9.63IAPKK17 pKa = 9.45TSFWRR22 pKa = 11.84IVRR25 pKa = 11.84VYY27 pKa = 10.54PSGQVQLDD35 pKa = 3.34QGMGRR40 pKa = 11.84SAYY43 pKa = 10.44LCPTDD48 pKa = 4.23SCLKK52 pKa = 10.42NALSKK57 pKa = 11.01NRR59 pKa = 11.84LGRR62 pKa = 11.84ALRR65 pKa = 11.84SPIPQDD71 pKa = 3.17LVQTLRR77 pKa = 11.84EE78 pKa = 3.93RR79 pKa = 11.84LINN82 pKa = 3.8
MM1 pKa = 8.03PINIRR6 pKa = 11.84RR7 pKa = 11.84CISCHH12 pKa = 6.11KK13 pKa = 9.63IAPKK17 pKa = 9.45TSFWRR22 pKa = 11.84IVRR25 pKa = 11.84VYY27 pKa = 10.54PSGQVQLDD35 pKa = 3.34QGMGRR40 pKa = 11.84SAYY43 pKa = 10.44LCPTDD48 pKa = 4.23SCLKK52 pKa = 10.42NALSKK57 pKa = 11.01NRR59 pKa = 11.84LGRR62 pKa = 11.84ALRR65 pKa = 11.84SPIPQDD71 pKa = 3.17LVQTLRR77 pKa = 11.84EE78 pKa = 3.93RR79 pKa = 11.84LINN82 pKa = 3.8
Molecular weight: 9.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1539476 |
23 |
6380 |
318.9 |
35.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.011 ± 0.033 | 0.942 ± 0.014 |
4.993 ± 0.034 | 6.396 ± 0.047 |
4.087 ± 0.027 | 6.402 ± 0.053 |
1.754 ± 0.019 | 7.577 ± 0.033 |
5.314 ± 0.038 | 11.141 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.792 ± 0.019 | 4.851 ± 0.053 |
4.468 ± 0.024 | 5.337 ± 0.037 |
4.639 ± 0.032 | 6.577 ± 0.031 |
5.973 ± 0.051 | 5.993 ± 0.027 |
1.4 ± 0.016 | 3.354 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
