
Red clover powdery mildew-associated totivirus 6
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
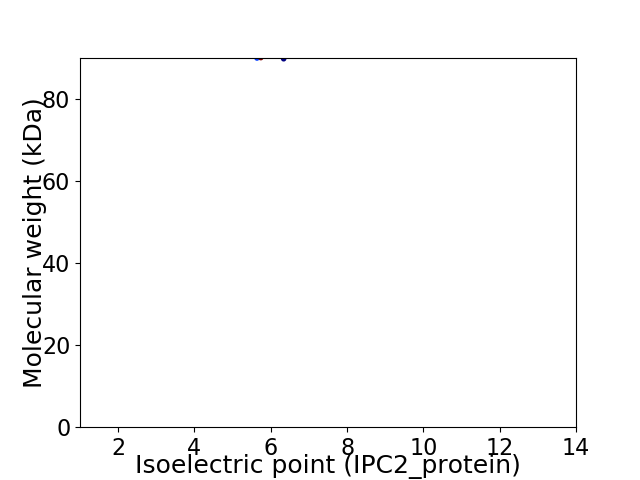
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
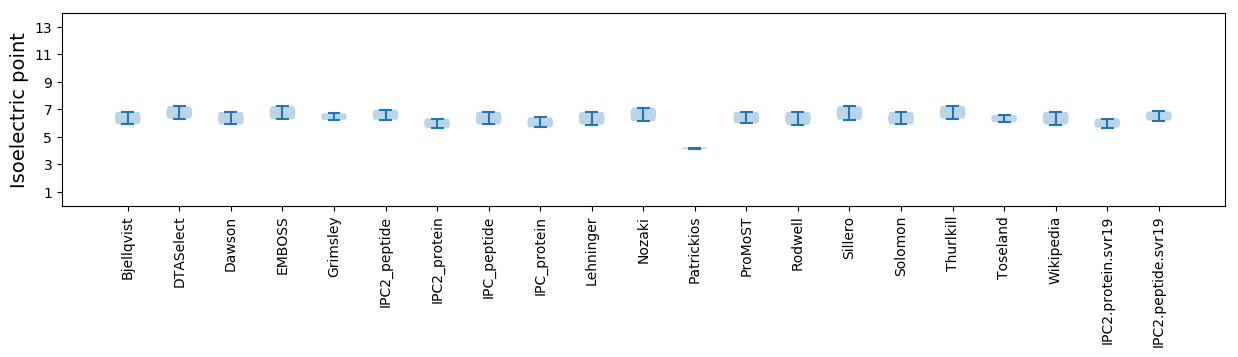
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S3Q2B4|A0A0S3Q2B4_9VIRU Capsid protein OS=Red clover powdery mildew-associated totivirus 6 OX=1714367 GN=CP PE=4 SV=1
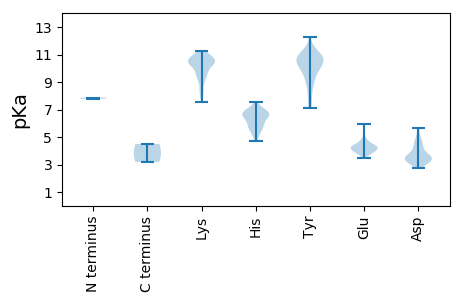
MM1 pKa = 7.79SFYY4 pKa = 11.0TLFNTSATFPKK15 pKa = 9.33MDD17 pKa = 3.43HH18 pKa = 6.51GKK20 pKa = 10.52FGMLNRR26 pKa = 11.84LNLRR30 pKa = 11.84LAAIEE35 pKa = 4.02KK36 pKa = 10.37KK37 pKa = 10.31KK38 pKa = 10.58ISEE41 pKa = 4.07AGYY44 pKa = 10.44DD45 pKa = 3.65GTRR48 pKa = 11.84EE49 pKa = 3.87EE50 pKa = 5.45AIAAPSGKK58 pKa = 8.73TSTLANEE65 pKa = 4.31ITDD68 pKa = 3.64EE69 pKa = 4.37LGFEE73 pKa = 4.12IDD75 pKa = 4.66HH76 pKa = 6.81RR77 pKa = 11.84APTNTLAEE85 pKa = 4.08GTIYY89 pKa = 10.58GRR91 pKa = 11.84NGTWRR96 pKa = 11.84ALGRR100 pKa = 11.84EE101 pKa = 4.04NTTSGLNPLSLNEE114 pKa = 4.58SGAIDD119 pKa = 4.13HH120 pKa = 6.58KK121 pKa = 10.12WLRR124 pKa = 11.84AQIRR128 pKa = 11.84EE129 pKa = 4.18LNPYY133 pKa = 10.02DD134 pKa = 3.9EE135 pKa = 4.54NRR137 pKa = 11.84EE138 pKa = 3.78ARR140 pKa = 11.84LNSFTTSWLTSVCYY154 pKa = 10.77DD155 pKa = 3.2NATALLLALGQRR167 pKa = 11.84LVIARR172 pKa = 11.84AAEE175 pKa = 3.84LAGTKK180 pKa = 8.08VVRR183 pKa = 11.84LVILEE188 pKa = 4.28SEE190 pKa = 4.49YY191 pKa = 11.02KK192 pKa = 10.86ANGLATLQNMVNHH205 pKa = 6.79ISEE208 pKa = 4.48KK209 pKa = 10.88LPTLFNGGIDD219 pKa = 3.61IANHH223 pKa = 7.07DD224 pKa = 4.65DD225 pKa = 5.26LISHH229 pKa = 7.11LAKK232 pKa = 10.92LLITKK237 pKa = 8.3FKK239 pKa = 10.51KK240 pKa = 10.48LPPLTEE246 pKa = 4.48AEE248 pKa = 4.49PKK250 pKa = 9.43RR251 pKa = 11.84AAYY254 pKa = 10.03EE255 pKa = 3.71DD256 pKa = 3.54VRR258 pKa = 11.84YY259 pKa = 9.63FEE261 pKa = 5.94CNEE264 pKa = 3.59FDD266 pKa = 4.61DD267 pKa = 4.21GHH269 pKa = 6.44SRR271 pKa = 11.84SGPTFGDD278 pKa = 3.32VYY280 pKa = 11.02GFSRR284 pKa = 11.84NRR286 pKa = 11.84ALVPLKK292 pKa = 10.92AFFAQYY298 pKa = 10.89GDD300 pKa = 3.34EE301 pKa = 4.8SVFKK305 pKa = 11.01DD306 pKa = 3.3EE307 pKa = 5.95GYY309 pKa = 11.2LSVQDD314 pKa = 4.43NEE316 pKa = 4.33HH317 pKa = 6.04EE318 pKa = 4.18QLFFNARR325 pKa = 11.84GHH327 pKa = 6.01VNLGTLTRR335 pKa = 11.84SEE337 pKa = 4.26IVFLDD342 pKa = 3.4SVIRR346 pKa = 11.84GNTRR350 pKa = 11.84QSPLMVDD357 pKa = 3.16QTFSLLPEE365 pKa = 4.76GSYY368 pKa = 11.4LMAYY372 pKa = 10.33NSTTTRR378 pKa = 11.84IEE380 pKa = 4.04DD381 pKa = 3.69EE382 pKa = 4.28AVVTSTLIRR391 pKa = 11.84SVIRR395 pKa = 11.84KK396 pKa = 8.45LVNNHH401 pKa = 5.12NWYY404 pKa = 10.35EE405 pKa = 4.1DD406 pKa = 3.17ARR408 pKa = 11.84TANLLLQQWSAQPATEE424 pKa = 4.17SVEE427 pKa = 4.42SHH429 pKa = 5.36WWPSLEE435 pKa = 3.93RR436 pKa = 11.84DD437 pKa = 3.61LRR439 pKa = 11.84VPTLGLGRR447 pKa = 11.84AAIPGLIGKK456 pKa = 9.8YY457 pKa = 9.48GVNVSKK463 pKa = 10.68QALIEE468 pKa = 4.34GQDD471 pKa = 3.66LYY473 pKa = 11.76GDD475 pKa = 4.46DD476 pKa = 4.08DD477 pKa = 5.02SKK479 pKa = 11.19FVKK482 pKa = 10.57GLIFNTLWYY491 pKa = 8.16WGEE494 pKa = 3.97YY495 pKa = 7.59LTIMNSSTMKK505 pKa = 10.51QLADD509 pKa = 3.38KK510 pKa = 10.91LRR512 pKa = 11.84FGGKK516 pKa = 8.17EE517 pKa = 3.6WLSEE521 pKa = 3.89QDD523 pKa = 2.76RR524 pKa = 11.84SYY526 pKa = 12.24AMISAISGMAVDD538 pKa = 5.54APASNISSTSIVGGIEE554 pKa = 3.72SQLRR558 pKa = 11.84NRR560 pKa = 11.84VSFGYY565 pKa = 9.75IDD567 pKa = 3.3TSSIVRR573 pKa = 11.84HH574 pKa = 6.31GYY576 pKa = 9.32SVSGSSLTYY585 pKa = 10.56NRR587 pKa = 11.84LVQPGSTCLIVGLNGTLIRR606 pKa = 11.84STPYY610 pKa = 10.66SSVFCLGAASSVDD623 pKa = 3.74VKK625 pKa = 10.57WRR627 pKa = 11.84TTMAYY632 pKa = 8.93TYY634 pKa = 11.17NDD636 pKa = 2.94MWALGVVARR645 pKa = 11.84WQGYY649 pKa = 9.51DD650 pKa = 2.78VDD652 pKa = 4.76YY653 pKa = 11.04RR654 pKa = 11.84LPDD657 pKa = 3.19QSEE660 pKa = 4.28TSNRR664 pKa = 11.84LHH666 pKa = 6.92AANDD670 pKa = 3.46SSVAYY675 pKa = 9.02PPLVPTLAQRR685 pKa = 11.84RR686 pKa = 11.84GFYY689 pKa = 10.05PIVDD693 pKa = 3.66IKK695 pKa = 10.51PRR697 pKa = 11.84KK698 pKa = 9.11RR699 pKa = 11.84CFGTDD704 pKa = 2.89TASLHH709 pKa = 5.41TYY711 pKa = 8.71KK712 pKa = 9.73RR713 pKa = 11.84TYY715 pKa = 9.33VWTRR719 pKa = 11.84DD720 pKa = 3.23TPTALEE726 pKa = 3.88QCRR729 pKa = 11.84LDD731 pKa = 3.42ARR733 pKa = 11.84LVGSEE738 pKa = 4.35PPSAAHH744 pKa = 6.93ALMVDD749 pKa = 3.52STTIATTSKK758 pKa = 10.4KK759 pKa = 10.36YY760 pKa = 10.69YY761 pKa = 9.63IRR763 pKa = 11.84SDD765 pKa = 3.4YY766 pKa = 10.47DD767 pKa = 3.47LSMSGFHH774 pKa = 4.98VTEE777 pKa = 4.34LATAITLPDD786 pKa = 3.17ATVALEE792 pKa = 4.05STRR795 pKa = 11.84LEE797 pKa = 4.65SIDD800 pKa = 4.46DD801 pKa = 4.02LPVDD805 pKa = 3.33QHH807 pKa = 6.52EE808 pKa = 4.2NN809 pKa = 3.19
MM1 pKa = 7.79SFYY4 pKa = 11.0TLFNTSATFPKK15 pKa = 9.33MDD17 pKa = 3.43HH18 pKa = 6.51GKK20 pKa = 10.52FGMLNRR26 pKa = 11.84LNLRR30 pKa = 11.84LAAIEE35 pKa = 4.02KK36 pKa = 10.37KK37 pKa = 10.31KK38 pKa = 10.58ISEE41 pKa = 4.07AGYY44 pKa = 10.44DD45 pKa = 3.65GTRR48 pKa = 11.84EE49 pKa = 3.87EE50 pKa = 5.45AIAAPSGKK58 pKa = 8.73TSTLANEE65 pKa = 4.31ITDD68 pKa = 3.64EE69 pKa = 4.37LGFEE73 pKa = 4.12IDD75 pKa = 4.66HH76 pKa = 6.81RR77 pKa = 11.84APTNTLAEE85 pKa = 4.08GTIYY89 pKa = 10.58GRR91 pKa = 11.84NGTWRR96 pKa = 11.84ALGRR100 pKa = 11.84EE101 pKa = 4.04NTTSGLNPLSLNEE114 pKa = 4.58SGAIDD119 pKa = 4.13HH120 pKa = 6.58KK121 pKa = 10.12WLRR124 pKa = 11.84AQIRR128 pKa = 11.84EE129 pKa = 4.18LNPYY133 pKa = 10.02DD134 pKa = 3.9EE135 pKa = 4.54NRR137 pKa = 11.84EE138 pKa = 3.78ARR140 pKa = 11.84LNSFTTSWLTSVCYY154 pKa = 10.77DD155 pKa = 3.2NATALLLALGQRR167 pKa = 11.84LVIARR172 pKa = 11.84AAEE175 pKa = 3.84LAGTKK180 pKa = 8.08VVRR183 pKa = 11.84LVILEE188 pKa = 4.28SEE190 pKa = 4.49YY191 pKa = 11.02KK192 pKa = 10.86ANGLATLQNMVNHH205 pKa = 6.79ISEE208 pKa = 4.48KK209 pKa = 10.88LPTLFNGGIDD219 pKa = 3.61IANHH223 pKa = 7.07DD224 pKa = 4.65DD225 pKa = 5.26LISHH229 pKa = 7.11LAKK232 pKa = 10.92LLITKK237 pKa = 8.3FKK239 pKa = 10.51KK240 pKa = 10.48LPPLTEE246 pKa = 4.48AEE248 pKa = 4.49PKK250 pKa = 9.43RR251 pKa = 11.84AAYY254 pKa = 10.03EE255 pKa = 3.71DD256 pKa = 3.54VRR258 pKa = 11.84YY259 pKa = 9.63FEE261 pKa = 5.94CNEE264 pKa = 3.59FDD266 pKa = 4.61DD267 pKa = 4.21GHH269 pKa = 6.44SRR271 pKa = 11.84SGPTFGDD278 pKa = 3.32VYY280 pKa = 11.02GFSRR284 pKa = 11.84NRR286 pKa = 11.84ALVPLKK292 pKa = 10.92AFFAQYY298 pKa = 10.89GDD300 pKa = 3.34EE301 pKa = 4.8SVFKK305 pKa = 11.01DD306 pKa = 3.3EE307 pKa = 5.95GYY309 pKa = 11.2LSVQDD314 pKa = 4.43NEE316 pKa = 4.33HH317 pKa = 6.04EE318 pKa = 4.18QLFFNARR325 pKa = 11.84GHH327 pKa = 6.01VNLGTLTRR335 pKa = 11.84SEE337 pKa = 4.26IVFLDD342 pKa = 3.4SVIRR346 pKa = 11.84GNTRR350 pKa = 11.84QSPLMVDD357 pKa = 3.16QTFSLLPEE365 pKa = 4.76GSYY368 pKa = 11.4LMAYY372 pKa = 10.33NSTTTRR378 pKa = 11.84IEE380 pKa = 4.04DD381 pKa = 3.69EE382 pKa = 4.28AVVTSTLIRR391 pKa = 11.84SVIRR395 pKa = 11.84KK396 pKa = 8.45LVNNHH401 pKa = 5.12NWYY404 pKa = 10.35EE405 pKa = 4.1DD406 pKa = 3.17ARR408 pKa = 11.84TANLLLQQWSAQPATEE424 pKa = 4.17SVEE427 pKa = 4.42SHH429 pKa = 5.36WWPSLEE435 pKa = 3.93RR436 pKa = 11.84DD437 pKa = 3.61LRR439 pKa = 11.84VPTLGLGRR447 pKa = 11.84AAIPGLIGKK456 pKa = 9.8YY457 pKa = 9.48GVNVSKK463 pKa = 10.68QALIEE468 pKa = 4.34GQDD471 pKa = 3.66LYY473 pKa = 11.76GDD475 pKa = 4.46DD476 pKa = 4.08DD477 pKa = 5.02SKK479 pKa = 11.19FVKK482 pKa = 10.57GLIFNTLWYY491 pKa = 8.16WGEE494 pKa = 3.97YY495 pKa = 7.59LTIMNSSTMKK505 pKa = 10.51QLADD509 pKa = 3.38KK510 pKa = 10.91LRR512 pKa = 11.84FGGKK516 pKa = 8.17EE517 pKa = 3.6WLSEE521 pKa = 3.89QDD523 pKa = 2.76RR524 pKa = 11.84SYY526 pKa = 12.24AMISAISGMAVDD538 pKa = 5.54APASNISSTSIVGGIEE554 pKa = 3.72SQLRR558 pKa = 11.84NRR560 pKa = 11.84VSFGYY565 pKa = 9.75IDD567 pKa = 3.3TSSIVRR573 pKa = 11.84HH574 pKa = 6.31GYY576 pKa = 9.32SVSGSSLTYY585 pKa = 10.56NRR587 pKa = 11.84LVQPGSTCLIVGLNGTLIRR606 pKa = 11.84STPYY610 pKa = 10.66SSVFCLGAASSVDD623 pKa = 3.74VKK625 pKa = 10.57WRR627 pKa = 11.84TTMAYY632 pKa = 8.93TYY634 pKa = 11.17NDD636 pKa = 2.94MWALGVVARR645 pKa = 11.84WQGYY649 pKa = 9.51DD650 pKa = 2.78VDD652 pKa = 4.76YY653 pKa = 11.04RR654 pKa = 11.84LPDD657 pKa = 3.19QSEE660 pKa = 4.28TSNRR664 pKa = 11.84LHH666 pKa = 6.92AANDD670 pKa = 3.46SSVAYY675 pKa = 9.02PPLVPTLAQRR685 pKa = 11.84RR686 pKa = 11.84GFYY689 pKa = 10.05PIVDD693 pKa = 3.66IKK695 pKa = 10.51PRR697 pKa = 11.84KK698 pKa = 9.11RR699 pKa = 11.84CFGTDD704 pKa = 2.89TASLHH709 pKa = 5.41TYY711 pKa = 8.71KK712 pKa = 9.73RR713 pKa = 11.84TYY715 pKa = 9.33VWTRR719 pKa = 11.84DD720 pKa = 3.23TPTALEE726 pKa = 3.88QCRR729 pKa = 11.84LDD731 pKa = 3.42ARR733 pKa = 11.84LVGSEE738 pKa = 4.35PPSAAHH744 pKa = 6.93ALMVDD749 pKa = 3.52STTIATTSKK758 pKa = 10.4KK759 pKa = 10.36YY760 pKa = 10.69YY761 pKa = 9.63IRR763 pKa = 11.84SDD765 pKa = 3.4YY766 pKa = 10.47DD767 pKa = 3.47LSMSGFHH774 pKa = 4.98VTEE777 pKa = 4.34LATAITLPDD786 pKa = 3.17ATVALEE792 pKa = 4.05STRR795 pKa = 11.84LEE797 pKa = 4.65SIDD800 pKa = 4.46DD801 pKa = 4.02LPVDD805 pKa = 3.33QHH807 pKa = 6.52EE808 pKa = 4.2NN809 pKa = 3.19
Molecular weight: 90.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3Q2B4|A0A0S3Q2B4_9VIRU Capsid protein OS=Red clover powdery mildew-associated totivirus 6 OX=1714367 GN=CP PE=4 SV=1
MM1 pKa = 7.87DD2 pKa = 5.66GIARR6 pKa = 11.84STTFEE11 pKa = 3.87RR12 pKa = 11.84ASYY15 pKa = 10.36SLVEE19 pKa = 4.95IIPGIDD25 pKa = 2.75IRR27 pKa = 11.84GQFFCPTYY35 pKa = 10.23DD36 pKa = 3.02GAMRR40 pKa = 11.84VCSVYY45 pKa = 10.7YY46 pKa = 10.18RR47 pKa = 11.84DD48 pKa = 3.51LHH50 pKa = 5.67MSAMYY55 pKa = 10.22LSHH58 pKa = 7.56VEE60 pKa = 4.2SLPALLPAARR70 pKa = 11.84VAVSRR75 pKa = 11.84TQFGYY80 pKa = 10.32DD81 pKa = 3.99LCPLGCVSQKK91 pKa = 9.6EE92 pKa = 4.15HH93 pKa = 6.99LAFLTNYY100 pKa = 9.91SSEE103 pKa = 3.83DD104 pKa = 3.73RR105 pKa = 11.84YY106 pKa = 9.2TSRR109 pKa = 11.84HH110 pKa = 4.71FEE112 pKa = 4.0KK113 pKa = 10.76LSAVLAGRR121 pKa = 11.84IEE123 pKa = 4.26LPHH126 pKa = 6.48EE127 pKa = 4.51KK128 pKa = 10.51ASATHH133 pKa = 6.07LRR135 pKa = 11.84HH136 pKa = 6.51IVLADD141 pKa = 3.61IAAVMAEE148 pKa = 4.18RR149 pKa = 11.84QQGVPDD155 pKa = 4.78LGNVAYY161 pKa = 10.58LLEE164 pKa = 4.13NAIRR168 pKa = 11.84DD169 pKa = 3.53GWATEE174 pKa = 4.07TFVVGMILWVLALRR188 pKa = 11.84QPIRR192 pKa = 11.84GWVLGSKK199 pKa = 8.17WWLQAHH205 pKa = 6.72ANMKK209 pKa = 9.73EE210 pKa = 3.73YY211 pKa = 10.71HH212 pKa = 6.85DD213 pKa = 4.5FIKK216 pKa = 10.17TKK218 pKa = 10.66LSLRR222 pKa = 11.84AKK224 pKa = 10.22ALQNLVGVDD233 pKa = 3.93LSQLFEE239 pKa = 4.37VEE241 pKa = 4.22VLVNRR246 pKa = 11.84GMGQVDD252 pKa = 3.59WKK254 pKa = 11.08AEE256 pKa = 3.78RR257 pKa = 11.84LHH259 pKa = 6.5RR260 pKa = 11.84VRR262 pKa = 11.84PSLATFGVDD271 pKa = 4.25AIRR274 pKa = 11.84AEE276 pKa = 4.12ACKK279 pKa = 10.54LFTKK283 pKa = 10.48SKK285 pKa = 10.15QLGGKK290 pKa = 9.0LRR292 pKa = 11.84PRR294 pKa = 11.84DD295 pKa = 3.3WGSYY299 pKa = 7.67WDD301 pKa = 4.48TRR303 pKa = 11.84WMWAPTGAFFSQHH316 pKa = 6.66PEE318 pKa = 3.7DD319 pKa = 4.01KK320 pKa = 10.77AYY322 pKa = 10.8LSKK325 pKa = 11.11DD326 pKa = 3.22FSSRR330 pKa = 11.84SKK332 pKa = 10.66FYY334 pKa = 10.92ALSSMPDD341 pKa = 3.12YY342 pKa = 11.11GLEE345 pKa = 3.93HH346 pKa = 6.52FLGRR350 pKa = 11.84RR351 pKa = 11.84CEE353 pKa = 4.11IQAKK357 pKa = 10.09ASQKK361 pKa = 9.77YY362 pKa = 7.1EE363 pKa = 3.8WGKK366 pKa = 9.04MRR368 pKa = 11.84AIYY371 pKa = 8.53GTDD374 pKa = 2.86VTSFVLTNFCMGDD387 pKa = 3.43CEE389 pKa = 4.61EE390 pKa = 4.26FLEE393 pKa = 4.75PVFPLKK399 pKa = 10.4EE400 pKa = 3.89AATLSHH406 pKa = 5.73VKK408 pKa = 10.56ARR410 pKa = 11.84VKK412 pKa = 10.59DD413 pKa = 3.68VLNNGVPYY421 pKa = 10.83CFDD424 pKa = 3.78FEE426 pKa = 5.21DD427 pKa = 5.0FNSQHH432 pKa = 6.68SGDD435 pKa = 3.62AMEE438 pKa = 5.17AVLLAFGEE446 pKa = 4.57VCEE449 pKa = 5.71DD450 pKa = 3.54MMSHH454 pKa = 5.91EE455 pKa = 3.96QRR457 pKa = 11.84AAFEE461 pKa = 4.42WTCGSIGKK469 pKa = 7.52QTIHH473 pKa = 6.9SPDD476 pKa = 3.28GKK478 pKa = 10.51DD479 pKa = 3.11YY480 pKa = 7.48TTKK483 pKa = 9.39HH484 pKa = 5.52TLLSGWRR491 pKa = 11.84LTTFMNTVLNYY502 pKa = 9.83IYY504 pKa = 10.87VQLCSEE510 pKa = 4.67GEE512 pKa = 4.01LAATIHH518 pKa = 6.24TGDD521 pKa = 4.07DD522 pKa = 3.12VLAAVEE528 pKa = 4.02NLGAVATLQRR538 pKa = 11.84NAIEE542 pKa = 4.24HH543 pKa = 5.5NVRR546 pKa = 11.84FQRR549 pKa = 11.84SKK551 pKa = 11.26CFLGGLAEE559 pKa = 4.4FVRR562 pKa = 11.84VDD564 pKa = 3.49HH565 pKa = 7.0LSGTGAQYY573 pKa = 10.98LARR576 pKa = 11.84ACSSFVHH583 pKa = 6.72GPMDD587 pKa = 3.26MSIPFDD593 pKa = 3.71PLAQVEE599 pKa = 4.27AMHH602 pKa = 6.43TRR604 pKa = 11.84TLEE607 pKa = 4.17CKK609 pKa = 9.96EE610 pKa = 4.0RR611 pKa = 11.84GADD614 pKa = 3.67VEE616 pKa = 4.69LMNALEE622 pKa = 4.29RR623 pKa = 11.84RR624 pKa = 11.84AISYY628 pKa = 8.52VLQVTDD634 pKa = 4.21CEE636 pKa = 4.36NVNVDD641 pKa = 5.09DD642 pKa = 4.89ILNTHH647 pKa = 5.85VAYY650 pKa = 10.41GGMSRR655 pKa = 11.84EE656 pKa = 3.86IDD658 pKa = 3.2EE659 pKa = 4.45GAVANKK665 pKa = 9.44IEE667 pKa = 4.04RR668 pKa = 11.84RR669 pKa = 11.84AVVDD673 pKa = 4.43LLEE676 pKa = 4.7EE677 pKa = 4.39PPGPEE682 pKa = 3.46EE683 pKa = 4.98DD684 pKa = 3.88GSNKK688 pKa = 9.54ILPGCKK694 pKa = 9.31AYY696 pKa = 10.3ARR698 pKa = 11.84HH699 pKa = 5.76VARR702 pKa = 11.84TYY704 pKa = 11.02NLQEE708 pKa = 4.28HH709 pKa = 5.86EE710 pKa = 4.24LKK712 pKa = 10.21IRR714 pKa = 11.84RR715 pKa = 11.84RR716 pKa = 11.84VQQALDD722 pKa = 3.4ASVTKK727 pKa = 9.43TKK729 pKa = 10.74YY730 pKa = 10.38AIRR733 pKa = 11.84VVKK736 pKa = 10.17NVSSISQSIARR747 pKa = 11.84ISQFKK752 pKa = 10.39KK753 pKa = 9.79FANIKK758 pKa = 10.45LGMKK762 pKa = 10.41ANLAKK767 pKa = 10.8AFGVPLTTGDD777 pKa = 3.26STQDD781 pKa = 2.87VKK783 pKa = 10.57TAMLISSEE791 pKa = 4.2DD792 pKa = 3.15QLAALRR798 pKa = 11.84LWFF801 pKa = 4.49
MM1 pKa = 7.87DD2 pKa = 5.66GIARR6 pKa = 11.84STTFEE11 pKa = 3.87RR12 pKa = 11.84ASYY15 pKa = 10.36SLVEE19 pKa = 4.95IIPGIDD25 pKa = 2.75IRR27 pKa = 11.84GQFFCPTYY35 pKa = 10.23DD36 pKa = 3.02GAMRR40 pKa = 11.84VCSVYY45 pKa = 10.7YY46 pKa = 10.18RR47 pKa = 11.84DD48 pKa = 3.51LHH50 pKa = 5.67MSAMYY55 pKa = 10.22LSHH58 pKa = 7.56VEE60 pKa = 4.2SLPALLPAARR70 pKa = 11.84VAVSRR75 pKa = 11.84TQFGYY80 pKa = 10.32DD81 pKa = 3.99LCPLGCVSQKK91 pKa = 9.6EE92 pKa = 4.15HH93 pKa = 6.99LAFLTNYY100 pKa = 9.91SSEE103 pKa = 3.83DD104 pKa = 3.73RR105 pKa = 11.84YY106 pKa = 9.2TSRR109 pKa = 11.84HH110 pKa = 4.71FEE112 pKa = 4.0KK113 pKa = 10.76LSAVLAGRR121 pKa = 11.84IEE123 pKa = 4.26LPHH126 pKa = 6.48EE127 pKa = 4.51KK128 pKa = 10.51ASATHH133 pKa = 6.07LRR135 pKa = 11.84HH136 pKa = 6.51IVLADD141 pKa = 3.61IAAVMAEE148 pKa = 4.18RR149 pKa = 11.84QQGVPDD155 pKa = 4.78LGNVAYY161 pKa = 10.58LLEE164 pKa = 4.13NAIRR168 pKa = 11.84DD169 pKa = 3.53GWATEE174 pKa = 4.07TFVVGMILWVLALRR188 pKa = 11.84QPIRR192 pKa = 11.84GWVLGSKK199 pKa = 8.17WWLQAHH205 pKa = 6.72ANMKK209 pKa = 9.73EE210 pKa = 3.73YY211 pKa = 10.71HH212 pKa = 6.85DD213 pKa = 4.5FIKK216 pKa = 10.17TKK218 pKa = 10.66LSLRR222 pKa = 11.84AKK224 pKa = 10.22ALQNLVGVDD233 pKa = 3.93LSQLFEE239 pKa = 4.37VEE241 pKa = 4.22VLVNRR246 pKa = 11.84GMGQVDD252 pKa = 3.59WKK254 pKa = 11.08AEE256 pKa = 3.78RR257 pKa = 11.84LHH259 pKa = 6.5RR260 pKa = 11.84VRR262 pKa = 11.84PSLATFGVDD271 pKa = 4.25AIRR274 pKa = 11.84AEE276 pKa = 4.12ACKK279 pKa = 10.54LFTKK283 pKa = 10.48SKK285 pKa = 10.15QLGGKK290 pKa = 9.0LRR292 pKa = 11.84PRR294 pKa = 11.84DD295 pKa = 3.3WGSYY299 pKa = 7.67WDD301 pKa = 4.48TRR303 pKa = 11.84WMWAPTGAFFSQHH316 pKa = 6.66PEE318 pKa = 3.7DD319 pKa = 4.01KK320 pKa = 10.77AYY322 pKa = 10.8LSKK325 pKa = 11.11DD326 pKa = 3.22FSSRR330 pKa = 11.84SKK332 pKa = 10.66FYY334 pKa = 10.92ALSSMPDD341 pKa = 3.12YY342 pKa = 11.11GLEE345 pKa = 3.93HH346 pKa = 6.52FLGRR350 pKa = 11.84RR351 pKa = 11.84CEE353 pKa = 4.11IQAKK357 pKa = 10.09ASQKK361 pKa = 9.77YY362 pKa = 7.1EE363 pKa = 3.8WGKK366 pKa = 9.04MRR368 pKa = 11.84AIYY371 pKa = 8.53GTDD374 pKa = 2.86VTSFVLTNFCMGDD387 pKa = 3.43CEE389 pKa = 4.61EE390 pKa = 4.26FLEE393 pKa = 4.75PVFPLKK399 pKa = 10.4EE400 pKa = 3.89AATLSHH406 pKa = 5.73VKK408 pKa = 10.56ARR410 pKa = 11.84VKK412 pKa = 10.59DD413 pKa = 3.68VLNNGVPYY421 pKa = 10.83CFDD424 pKa = 3.78FEE426 pKa = 5.21DD427 pKa = 5.0FNSQHH432 pKa = 6.68SGDD435 pKa = 3.62AMEE438 pKa = 5.17AVLLAFGEE446 pKa = 4.57VCEE449 pKa = 5.71DD450 pKa = 3.54MMSHH454 pKa = 5.91EE455 pKa = 3.96QRR457 pKa = 11.84AAFEE461 pKa = 4.42WTCGSIGKK469 pKa = 7.52QTIHH473 pKa = 6.9SPDD476 pKa = 3.28GKK478 pKa = 10.51DD479 pKa = 3.11YY480 pKa = 7.48TTKK483 pKa = 9.39HH484 pKa = 5.52TLLSGWRR491 pKa = 11.84LTTFMNTVLNYY502 pKa = 9.83IYY504 pKa = 10.87VQLCSEE510 pKa = 4.67GEE512 pKa = 4.01LAATIHH518 pKa = 6.24TGDD521 pKa = 4.07DD522 pKa = 3.12VLAAVEE528 pKa = 4.02NLGAVATLQRR538 pKa = 11.84NAIEE542 pKa = 4.24HH543 pKa = 5.5NVRR546 pKa = 11.84FQRR549 pKa = 11.84SKK551 pKa = 11.26CFLGGLAEE559 pKa = 4.4FVRR562 pKa = 11.84VDD564 pKa = 3.49HH565 pKa = 7.0LSGTGAQYY573 pKa = 10.98LARR576 pKa = 11.84ACSSFVHH583 pKa = 6.72GPMDD587 pKa = 3.26MSIPFDD593 pKa = 3.71PLAQVEE599 pKa = 4.27AMHH602 pKa = 6.43TRR604 pKa = 11.84TLEE607 pKa = 4.17CKK609 pKa = 9.96EE610 pKa = 4.0RR611 pKa = 11.84GADD614 pKa = 3.67VEE616 pKa = 4.69LMNALEE622 pKa = 4.29RR623 pKa = 11.84RR624 pKa = 11.84AISYY628 pKa = 8.52VLQVTDD634 pKa = 4.21CEE636 pKa = 4.36NVNVDD641 pKa = 5.09DD642 pKa = 4.89ILNTHH647 pKa = 5.85VAYY650 pKa = 10.41GGMSRR655 pKa = 11.84EE656 pKa = 3.86IDD658 pKa = 3.2EE659 pKa = 4.45GAVANKK665 pKa = 9.44IEE667 pKa = 4.04RR668 pKa = 11.84RR669 pKa = 11.84AVVDD673 pKa = 4.43LLEE676 pKa = 4.7EE677 pKa = 4.39PPGPEE682 pKa = 3.46EE683 pKa = 4.98DD684 pKa = 3.88GSNKK688 pKa = 9.54ILPGCKK694 pKa = 9.31AYY696 pKa = 10.3ARR698 pKa = 11.84HH699 pKa = 5.76VARR702 pKa = 11.84TYY704 pKa = 11.02NLQEE708 pKa = 4.28HH709 pKa = 5.86EE710 pKa = 4.24LKK712 pKa = 10.21IRR714 pKa = 11.84RR715 pKa = 11.84RR716 pKa = 11.84VQQALDD722 pKa = 3.4ASVTKK727 pKa = 9.43TKK729 pKa = 10.74YY730 pKa = 10.38AIRR733 pKa = 11.84VVKK736 pKa = 10.17NVSSISQSIARR747 pKa = 11.84ISQFKK752 pKa = 10.39KK753 pKa = 9.79FANIKK758 pKa = 10.45LGMKK762 pKa = 10.41ANLAKK767 pKa = 10.8AFGVPLTTGDD777 pKa = 3.26STQDD781 pKa = 2.87VKK783 pKa = 10.57TAMLISSEE791 pKa = 4.2DD792 pKa = 3.15QLAALRR798 pKa = 11.84LWFF801 pKa = 4.49
Molecular weight: 89.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1610 |
801 |
809 |
805.0 |
89.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.068 ± 0.562 | 1.429 ± 0.49 |
5.59 ± 0.157 | 6.025 ± 0.242 |
3.851 ± 0.367 | 6.522 ± 0.109 |
2.609 ± 0.362 | 4.534 ± 0.293 |
4.41 ± 0.413 | 10.186 ± 0.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.405 | 4.037 ± 0.648 |
3.478 ± 0.252 | 3.23 ± 0.364 |
6.335 ± 0.022 | 7.764 ± 0.723 |
6.584 ± 1.036 | 6.584 ± 0.641 |
1.739 ± 0.006 | 3.727 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
