
Synechococcus phage ACG-2014d
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
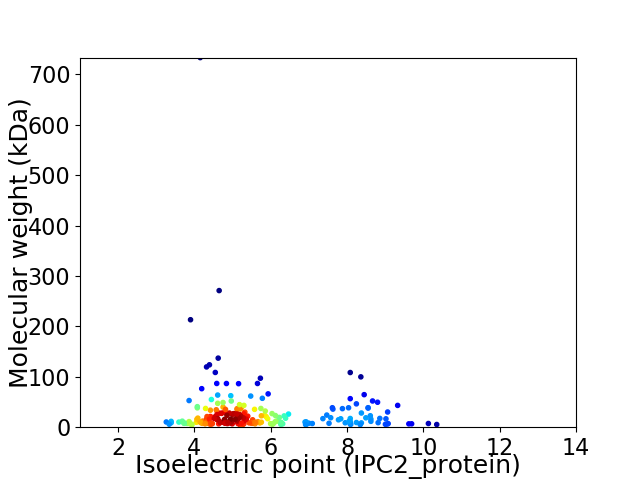
Virtual 2D-PAGE plot for 218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3HXY0|A0A0E3HXY0_9CAUD Uncharacterized protein OS=Synechococcus phage ACG-2014d OX=1493509 GN=Syn7803C102_172 PE=4 SV=1
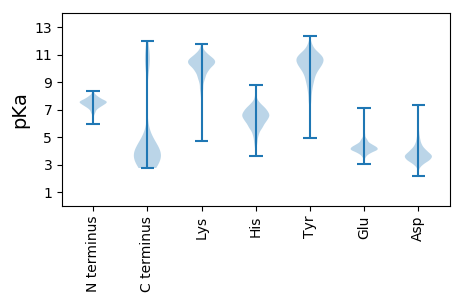
MM1 pKa = 7.17ATNTGALANIKK12 pKa = 9.65PSSGTFTTLFKK23 pKa = 11.22NDD25 pKa = 3.64VLSSTTGTVFVNCDD39 pKa = 2.66GTGADD44 pKa = 3.89TYY46 pKa = 11.17NIKK49 pKa = 10.6LNRR52 pKa = 11.84WDD54 pKa = 4.1QEE56 pKa = 4.02LTLDD60 pKa = 3.57ANTYY64 pKa = 9.72LLHH67 pKa = 7.15RR68 pKa = 11.84GDD70 pKa = 5.33LISNVKK76 pKa = 7.51WTLSASIPLADD87 pKa = 6.09AIPGTKK93 pKa = 9.46FVSSDD98 pKa = 3.67GEE100 pKa = 4.02KK101 pKa = 10.29CAYY104 pKa = 9.95LLDD107 pKa = 3.96VADD110 pKa = 4.78PVTTTYY116 pKa = 10.21EE117 pKa = 3.71VRR119 pKa = 11.84YY120 pKa = 9.43KK121 pKa = 11.01SLIAFTLEE129 pKa = 3.81NVADD133 pKa = 4.03TGSSVNPDD141 pKa = 3.38YY142 pKa = 11.88ANGEE146 pKa = 4.45TVSNGGGVSGVVYY159 pKa = 10.38EE160 pKa = 5.1NIPGSNDD167 pKa = 4.12DD168 pKa = 3.56AVLWIGDD175 pKa = 3.45ITGGTFSEE183 pKa = 4.66GDD185 pKa = 3.75VLTGGSSTTSGTVSTGGIATAANRR209 pKa = 11.84FVFNDD214 pKa = 4.0GAGGAVFRR222 pKa = 11.84LQNEE226 pKa = 4.16IQPEE230 pKa = 4.26LLTDD234 pKa = 3.84RR235 pKa = 11.84VYY237 pKa = 11.3KK238 pKa = 10.56FDD240 pKa = 3.52VANASMSGKK249 pKa = 9.52ILEE252 pKa = 4.63FSDD255 pKa = 4.04TNGGSNNSGDD265 pKa = 3.66EE266 pKa = 4.08FVTGKK271 pKa = 8.13TVSGTPGQAGAFVQYY286 pKa = 10.86NLTGAEE292 pKa = 4.7LISNFYY298 pKa = 10.55PYY300 pKa = 10.79DD301 pKa = 3.62QADD304 pKa = 3.22ATYY307 pKa = 10.9ADD309 pKa = 4.08DD310 pKa = 3.61SQYY313 pKa = 9.75FTFSEE318 pKa = 4.9EE319 pKa = 4.04YY320 pKa = 8.93TFNEE324 pKa = 4.19IYY326 pKa = 10.77VYY328 pKa = 7.45TQKK331 pKa = 11.12DD332 pKa = 3.95EE333 pKa = 4.22YY334 pKa = 11.06LQTPDD339 pKa = 2.56QWIITDD345 pKa = 3.54AFIYY349 pKa = 10.43RR350 pKa = 11.84DD351 pKa = 3.21VTYY354 pKa = 10.67AVDD357 pKa = 5.39AIAGDD362 pKa = 3.72SYY364 pKa = 11.3GTILEE369 pKa = 4.11WDD371 pKa = 3.59KK372 pKa = 11.7VNSKK376 pKa = 10.82LYY378 pKa = 9.23VANGPGSAAWAGSDD392 pKa = 3.49TFFEE396 pKa = 4.73SPRR399 pKa = 11.84QLSVSKK405 pKa = 9.88ATATINSVLSSPADD419 pKa = 3.68DD420 pKa = 4.62FIVRR424 pKa = 11.84ADD426 pKa = 4.84AIAQEE431 pKa = 4.32TTEE434 pKa = 4.23RR435 pKa = 11.84QTGIIIGPGQSIQVEE450 pKa = 4.64CTNGRR455 pKa = 11.84CNFVFDD461 pKa = 5.03AFQDD465 pKa = 3.86TVNEE469 pKa = 4.35VVTALYY475 pKa = 10.07QRR477 pKa = 11.84SSDD480 pKa = 3.82YY481 pKa = 9.65QTGLEE486 pKa = 4.1DD487 pKa = 4.62SGDD490 pKa = 3.85GGDD493 pKa = 5.04GGDD496 pKa = 3.35
MM1 pKa = 7.17ATNTGALANIKK12 pKa = 9.65PSSGTFTTLFKK23 pKa = 11.22NDD25 pKa = 3.64VLSSTTGTVFVNCDD39 pKa = 2.66GTGADD44 pKa = 3.89TYY46 pKa = 11.17NIKK49 pKa = 10.6LNRR52 pKa = 11.84WDD54 pKa = 4.1QEE56 pKa = 4.02LTLDD60 pKa = 3.57ANTYY64 pKa = 9.72LLHH67 pKa = 7.15RR68 pKa = 11.84GDD70 pKa = 5.33LISNVKK76 pKa = 7.51WTLSASIPLADD87 pKa = 6.09AIPGTKK93 pKa = 9.46FVSSDD98 pKa = 3.67GEE100 pKa = 4.02KK101 pKa = 10.29CAYY104 pKa = 9.95LLDD107 pKa = 3.96VADD110 pKa = 4.78PVTTTYY116 pKa = 10.21EE117 pKa = 3.71VRR119 pKa = 11.84YY120 pKa = 9.43KK121 pKa = 11.01SLIAFTLEE129 pKa = 3.81NVADD133 pKa = 4.03TGSSVNPDD141 pKa = 3.38YY142 pKa = 11.88ANGEE146 pKa = 4.45TVSNGGGVSGVVYY159 pKa = 10.38EE160 pKa = 5.1NIPGSNDD167 pKa = 4.12DD168 pKa = 3.56AVLWIGDD175 pKa = 3.45ITGGTFSEE183 pKa = 4.66GDD185 pKa = 3.75VLTGGSSTTSGTVSTGGIATAANRR209 pKa = 11.84FVFNDD214 pKa = 4.0GAGGAVFRR222 pKa = 11.84LQNEE226 pKa = 4.16IQPEE230 pKa = 4.26LLTDD234 pKa = 3.84RR235 pKa = 11.84VYY237 pKa = 11.3KK238 pKa = 10.56FDD240 pKa = 3.52VANASMSGKK249 pKa = 9.52ILEE252 pKa = 4.63FSDD255 pKa = 4.04TNGGSNNSGDD265 pKa = 3.66EE266 pKa = 4.08FVTGKK271 pKa = 8.13TVSGTPGQAGAFVQYY286 pKa = 10.86NLTGAEE292 pKa = 4.7LISNFYY298 pKa = 10.55PYY300 pKa = 10.79DD301 pKa = 3.62QADD304 pKa = 3.22ATYY307 pKa = 10.9ADD309 pKa = 4.08DD310 pKa = 3.61SQYY313 pKa = 9.75FTFSEE318 pKa = 4.9EE319 pKa = 4.04YY320 pKa = 8.93TFNEE324 pKa = 4.19IYY326 pKa = 10.77VYY328 pKa = 7.45TQKK331 pKa = 11.12DD332 pKa = 3.95EE333 pKa = 4.22YY334 pKa = 11.06LQTPDD339 pKa = 2.56QWIITDD345 pKa = 3.54AFIYY349 pKa = 10.43RR350 pKa = 11.84DD351 pKa = 3.21VTYY354 pKa = 10.67AVDD357 pKa = 5.39AIAGDD362 pKa = 3.72SYY364 pKa = 11.3GTILEE369 pKa = 4.11WDD371 pKa = 3.59KK372 pKa = 11.7VNSKK376 pKa = 10.82LYY378 pKa = 9.23VANGPGSAAWAGSDD392 pKa = 3.49TFFEE396 pKa = 4.73SPRR399 pKa = 11.84QLSVSKK405 pKa = 9.88ATATINSVLSSPADD419 pKa = 3.68DD420 pKa = 4.62FIVRR424 pKa = 11.84ADD426 pKa = 4.84AIAQEE431 pKa = 4.32TTEE434 pKa = 4.23RR435 pKa = 11.84QTGIIIGPGQSIQVEE450 pKa = 4.64CTNGRR455 pKa = 11.84CNFVFDD461 pKa = 5.03AFQDD465 pKa = 3.86TVNEE469 pKa = 4.35VVTALYY475 pKa = 10.07QRR477 pKa = 11.84SSDD480 pKa = 3.82YY481 pKa = 9.65QTGLEE486 pKa = 4.1DD487 pKa = 4.62SGDD490 pKa = 3.85GGDD493 pKa = 5.04GGDD496 pKa = 3.35
Molecular weight: 52.88 kDa
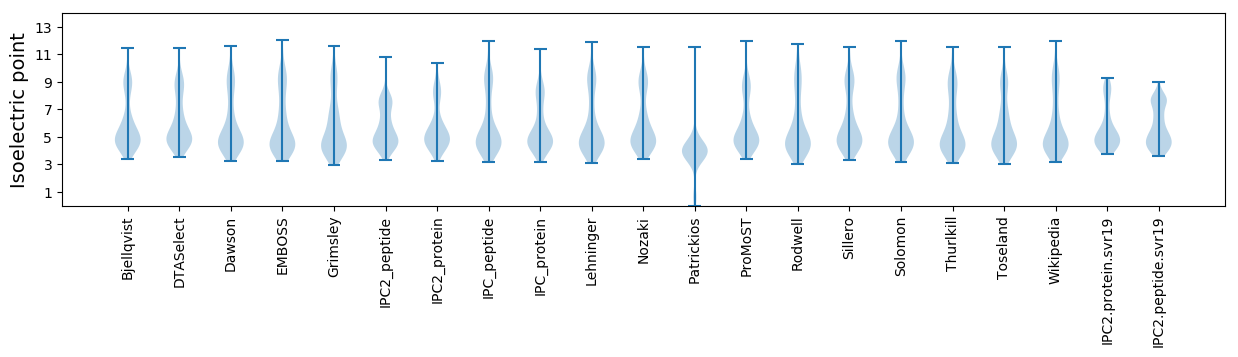
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3FUR1|A0A0E3FUR1_9CAUD Uncharacterized protein OS=Synechococcus phage ACG-2014d OX=1493509 GN=Syn7803C102_45 PE=4 SV=1
MM1 pKa = 7.56LSLVSIIGSSPVEE14 pKa = 3.67AKK16 pKa = 10.57GNKK19 pKa = 9.23LSGRR23 pKa = 11.84PGVSVRR29 pKa = 11.84PRR31 pKa = 11.84KK32 pKa = 10.13CNLCQPKK39 pKa = 9.69VRR41 pKa = 11.84IPKK44 pKa = 9.72KK45 pKa = 10.7CNLCQGGHH53 pKa = 6.2RR54 pKa = 11.84FGKK57 pKa = 9.01TKK59 pKa = 8.3MTFF62 pKa = 2.92
MM1 pKa = 7.56LSLVSIIGSSPVEE14 pKa = 3.67AKK16 pKa = 10.57GNKK19 pKa = 9.23LSGRR23 pKa = 11.84PGVSVRR29 pKa = 11.84PRR31 pKa = 11.84KK32 pKa = 10.13CNLCQPKK39 pKa = 9.69VRR41 pKa = 11.84IPKK44 pKa = 9.72KK45 pKa = 10.7CNLCQGGHH53 pKa = 6.2RR54 pKa = 11.84FGKK57 pKa = 9.01TKK59 pKa = 8.3MTFF62 pKa = 2.92
Molecular weight: 6.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
57254 |
35 |
6817 |
262.6 |
29.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.803 ± 0.204 | 0.973 ± 0.091 |
6.798 ± 0.133 | 6.141 ± 0.229 |
4.281 ± 0.107 | 7.813 ± 0.287 |
1.423 ± 0.109 | 6.351 ± 0.215 |
5.78 ± 0.447 | 7.166 ± 0.175 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.077 ± 0.212 | 5.879 ± 0.157 |
4.084 ± 0.145 | 3.745 ± 0.086 |
3.877 ± 0.158 | 7.421 ± 0.217 |
7.282 ± 0.39 | 6.642 ± 0.183 |
1.165 ± 0.095 | 4.297 ± 0.135 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
