
Micromonospora avicenniae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Micromonospora
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
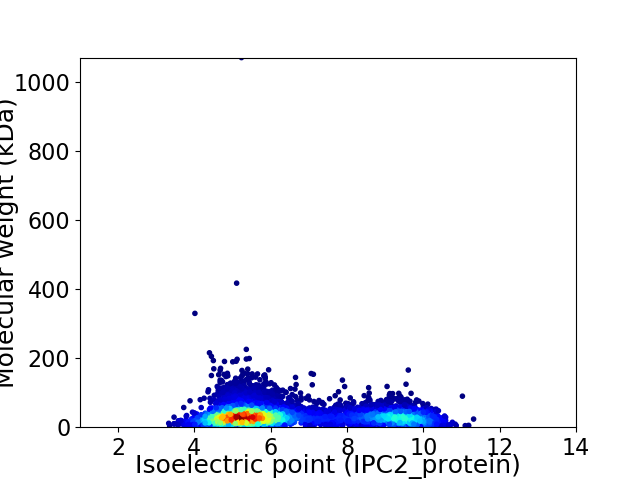
Virtual 2D-PAGE plot for 6031 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
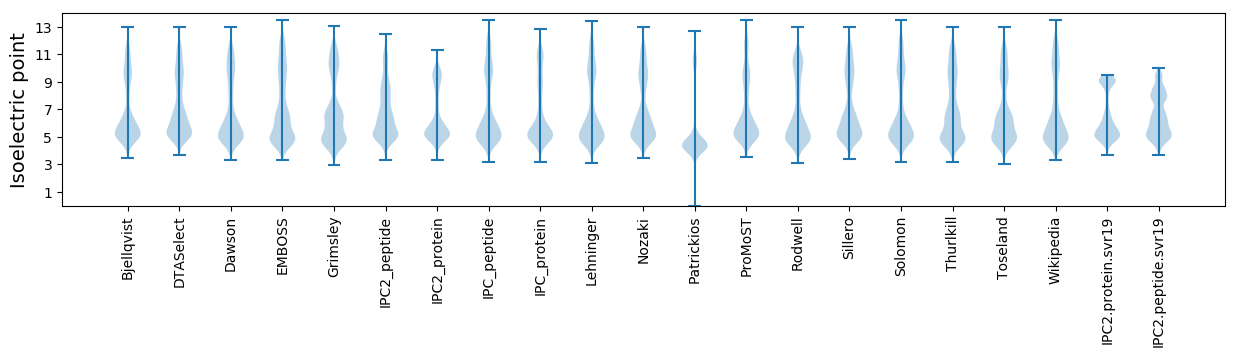
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N7B923|A0A1N7B923_9ACTN Undecaprenyl-diphosphatase OS=Micromonospora avicenniae OX=1198245 GN=SAMN05444858_110192 PE=4 SV=1
MM1 pKa = 7.16TKK3 pKa = 10.05VVYY6 pKa = 9.95AAEE9 pKa = 4.23PAVGWRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84LGAIPVVPVLVSALLPVLVLSSPAAAADD45 pKa = 3.65LNMTVRR51 pKa = 11.84VTVEE55 pKa = 3.61RR56 pKa = 11.84VEE58 pKa = 4.11GHH60 pKa = 7.24DD61 pKa = 3.97FGEE64 pKa = 4.81DD65 pKa = 3.3PDD67 pKa = 4.5FYY69 pKa = 11.26SYY71 pKa = 11.79VEE73 pKa = 3.83IAGEE77 pKa = 4.16SFNNEE82 pKa = 3.97DD83 pKa = 4.32TPASDD88 pKa = 4.02PFEE91 pKa = 4.32DD92 pKa = 4.56TYY94 pKa = 11.14TIEE97 pKa = 4.51PNWEE101 pKa = 3.65FSKK104 pKa = 10.89QVALSSGTVPIYY116 pKa = 10.92LEE118 pKa = 4.09IRR120 pKa = 11.84DD121 pKa = 3.68EE122 pKa = 4.7DD123 pKa = 3.74GFLRR127 pKa = 11.84FDD129 pKa = 5.46DD130 pKa = 5.47DD131 pKa = 5.68FADD134 pKa = 5.01LDD136 pKa = 4.53PDD138 pKa = 4.9GDD140 pKa = 4.72DD141 pKa = 5.09PDD143 pKa = 6.1DD144 pKa = 4.24PTDD147 pKa = 4.05NDD149 pKa = 4.47DD150 pKa = 5.38DD151 pKa = 4.37TDD153 pKa = 3.57QSLSFTVDD161 pKa = 3.45LNACTVSGDD170 pKa = 3.8LNGGCGVFTSTGTPDD185 pKa = 2.97EE186 pKa = 4.39SASIRR191 pKa = 11.84LRR193 pKa = 11.84VEE195 pKa = 3.7VTEE198 pKa = 4.66PPAAPGIRR206 pKa = 11.84IRR208 pKa = 11.84CMHH211 pKa = 6.8TPLFPQVGDD220 pKa = 3.72TVTITAEE227 pKa = 4.07FLDD230 pKa = 4.75GALTPARR237 pKa = 11.84ADD239 pKa = 3.52TLEE242 pKa = 3.46IWVNDD247 pKa = 3.31RR248 pKa = 11.84TAPDD252 pKa = 3.26VSTANSLTLTYY263 pKa = 10.96NHH265 pKa = 7.71TITGGGSFSYY275 pKa = 10.48GCRR278 pKa = 11.84GVDD281 pKa = 3.2DD282 pKa = 4.38SVPIFTGWRR291 pKa = 11.84SVAATAGAGDD301 pKa = 4.55EE302 pKa = 4.57VPVLYY307 pKa = 10.13TGSTYY312 pKa = 11.16SRR314 pKa = 11.84VDD316 pKa = 2.95WVFVGDD322 pKa = 3.71SDD324 pKa = 5.39DD325 pKa = 4.04YY326 pKa = 11.94NGAADD331 pKa = 4.22PQFQSDD337 pKa = 4.13VQQIIYY343 pKa = 10.73NSFYY347 pKa = 10.74SYY349 pKa = 11.61DD350 pKa = 3.79PFLVHH355 pKa = 6.46QDD357 pKa = 3.56AMNFWLQVGTGQADD371 pKa = 3.66PAEE374 pKa = 5.06DD375 pKa = 4.15GCDD378 pKa = 3.11HH379 pKa = 6.32VVPDD383 pKa = 4.52SGWADD388 pKa = 3.06SVMVVHH394 pKa = 6.63VDD396 pKa = 3.44SFRR399 pKa = 11.84DD400 pKa = 3.55CAMGGSNVFSGEE412 pKa = 3.84NTSFSTIRR420 pKa = 11.84HH421 pKa = 5.42EE422 pKa = 4.54SGHH425 pKa = 6.14RR426 pKa = 11.84PFGLADD432 pKa = 4.44EE433 pKa = 4.77YY434 pKa = 11.47CCDD437 pKa = 3.18GGYY440 pKa = 9.59YY441 pKa = 9.12QQDD444 pKa = 4.11VYY446 pKa = 11.29PNVYY450 pKa = 10.36DD451 pKa = 4.27EE452 pKa = 5.66PEE454 pKa = 4.56DD455 pKa = 4.2CADD458 pKa = 4.4DD459 pKa = 5.13APDD462 pKa = 4.11LGRR465 pKa = 11.84TASDD469 pKa = 3.04CRR471 pKa = 11.84EE472 pKa = 3.85FEE474 pKa = 4.23EE475 pKa = 6.0EE476 pKa = 4.09IEE478 pKa = 4.11NWDD481 pKa = 3.71DD482 pKa = 3.9FDD484 pKa = 4.42WSVSDD489 pKa = 4.18PATNDD494 pKa = 3.24LMVDD498 pKa = 3.25NTTAQAADD506 pKa = 3.38TRR508 pKa = 11.84RR509 pKa = 11.84IEE511 pKa = 3.94WYY513 pKa = 9.66FAEE516 pKa = 4.94CRR518 pKa = 11.84GAKK521 pKa = 9.57CC522 pKa = 4.55
MM1 pKa = 7.16TKK3 pKa = 10.05VVYY6 pKa = 9.95AAEE9 pKa = 4.23PAVGWRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84LGAIPVVPVLVSALLPVLVLSSPAAAADD45 pKa = 3.65LNMTVRR51 pKa = 11.84VTVEE55 pKa = 3.61RR56 pKa = 11.84VEE58 pKa = 4.11GHH60 pKa = 7.24DD61 pKa = 3.97FGEE64 pKa = 4.81DD65 pKa = 3.3PDD67 pKa = 4.5FYY69 pKa = 11.26SYY71 pKa = 11.79VEE73 pKa = 3.83IAGEE77 pKa = 4.16SFNNEE82 pKa = 3.97DD83 pKa = 4.32TPASDD88 pKa = 4.02PFEE91 pKa = 4.32DD92 pKa = 4.56TYY94 pKa = 11.14TIEE97 pKa = 4.51PNWEE101 pKa = 3.65FSKK104 pKa = 10.89QVALSSGTVPIYY116 pKa = 10.92LEE118 pKa = 4.09IRR120 pKa = 11.84DD121 pKa = 3.68EE122 pKa = 4.7DD123 pKa = 3.74GFLRR127 pKa = 11.84FDD129 pKa = 5.46DD130 pKa = 5.47DD131 pKa = 5.68FADD134 pKa = 5.01LDD136 pKa = 4.53PDD138 pKa = 4.9GDD140 pKa = 4.72DD141 pKa = 5.09PDD143 pKa = 6.1DD144 pKa = 4.24PTDD147 pKa = 4.05NDD149 pKa = 4.47DD150 pKa = 5.38DD151 pKa = 4.37TDD153 pKa = 3.57QSLSFTVDD161 pKa = 3.45LNACTVSGDD170 pKa = 3.8LNGGCGVFTSTGTPDD185 pKa = 2.97EE186 pKa = 4.39SASIRR191 pKa = 11.84LRR193 pKa = 11.84VEE195 pKa = 3.7VTEE198 pKa = 4.66PPAAPGIRR206 pKa = 11.84IRR208 pKa = 11.84CMHH211 pKa = 6.8TPLFPQVGDD220 pKa = 3.72TVTITAEE227 pKa = 4.07FLDD230 pKa = 4.75GALTPARR237 pKa = 11.84ADD239 pKa = 3.52TLEE242 pKa = 3.46IWVNDD247 pKa = 3.31RR248 pKa = 11.84TAPDD252 pKa = 3.26VSTANSLTLTYY263 pKa = 10.96NHH265 pKa = 7.71TITGGGSFSYY275 pKa = 10.48GCRR278 pKa = 11.84GVDD281 pKa = 3.2DD282 pKa = 4.38SVPIFTGWRR291 pKa = 11.84SVAATAGAGDD301 pKa = 4.55EE302 pKa = 4.57VPVLYY307 pKa = 10.13TGSTYY312 pKa = 11.16SRR314 pKa = 11.84VDD316 pKa = 2.95WVFVGDD322 pKa = 3.71SDD324 pKa = 5.39DD325 pKa = 4.04YY326 pKa = 11.94NGAADD331 pKa = 4.22PQFQSDD337 pKa = 4.13VQQIIYY343 pKa = 10.73NSFYY347 pKa = 10.74SYY349 pKa = 11.61DD350 pKa = 3.79PFLVHH355 pKa = 6.46QDD357 pKa = 3.56AMNFWLQVGTGQADD371 pKa = 3.66PAEE374 pKa = 5.06DD375 pKa = 4.15GCDD378 pKa = 3.11HH379 pKa = 6.32VVPDD383 pKa = 4.52SGWADD388 pKa = 3.06SVMVVHH394 pKa = 6.63VDD396 pKa = 3.44SFRR399 pKa = 11.84DD400 pKa = 3.55CAMGGSNVFSGEE412 pKa = 3.84NTSFSTIRR420 pKa = 11.84HH421 pKa = 5.42EE422 pKa = 4.54SGHH425 pKa = 6.14RR426 pKa = 11.84PFGLADD432 pKa = 4.44EE433 pKa = 4.77YY434 pKa = 11.47CCDD437 pKa = 3.18GGYY440 pKa = 9.59YY441 pKa = 9.12QQDD444 pKa = 4.11VYY446 pKa = 11.29PNVYY450 pKa = 10.36DD451 pKa = 4.27EE452 pKa = 5.66PEE454 pKa = 4.56DD455 pKa = 4.2CADD458 pKa = 4.4DD459 pKa = 5.13APDD462 pKa = 4.11LGRR465 pKa = 11.84TASDD469 pKa = 3.04CRR471 pKa = 11.84EE472 pKa = 3.85FEE474 pKa = 4.23EE475 pKa = 6.0EE476 pKa = 4.09IEE478 pKa = 4.11NWDD481 pKa = 3.71DD482 pKa = 3.9FDD484 pKa = 4.42WSVSDD489 pKa = 4.18PATNDD494 pKa = 3.24LMVDD498 pKa = 3.25NTTAQAADD506 pKa = 3.38TRR508 pKa = 11.84RR509 pKa = 11.84IEE511 pKa = 3.94WYY513 pKa = 9.66FAEE516 pKa = 4.94CRR518 pKa = 11.84GAKK521 pKa = 9.57CC522 pKa = 4.55
Molecular weight: 57.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N6R361|A0A1N6R361_9ACTN Dihydropteroate synthase OS=Micromonospora avicenniae OX=1198245 GN=SAMN05444858_101576 PE=3 SV=1
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84LGKK33 pKa = 10.04
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84LGKK33 pKa = 10.04
Molecular weight: 4.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1997338 |
24 |
10050 |
331.2 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.771 ± 0.048 | 0.681 ± 0.008 |
6.064 ± 0.028 | 5.21 ± 0.028 |
2.659 ± 0.016 | 9.403 ± 0.031 |
2.061 ± 0.014 | 3.266 ± 0.022 |
1.713 ± 0.026 | 10.414 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.625 ± 0.012 | 1.881 ± 0.022 |
6.391 ± 0.034 | 2.808 ± 0.019 |
8.382 ± 0.041 | 4.83 ± 0.026 |
6.117 ± 0.028 | 8.992 ± 0.033 |
1.589 ± 0.013 | 2.143 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
