
Eubacterium sp. CAG:115
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; environmental samples
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
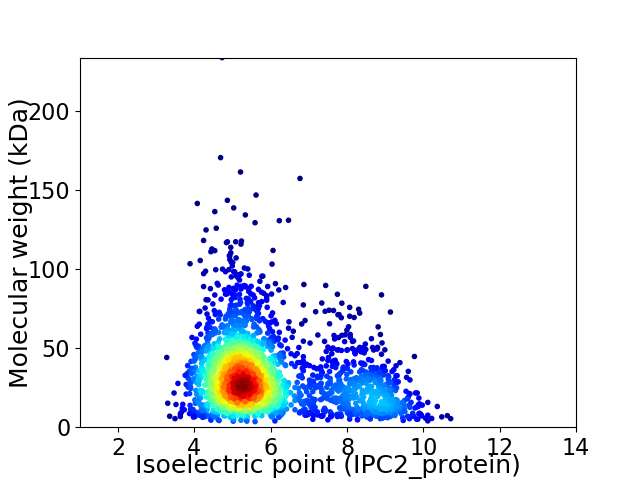
Virtual 2D-PAGE plot for 2289 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5L9S3|R5L9S3_9FIRM Uncharacterized protein OS=Eubacterium sp. CAG:115 OX=1262878 GN=BN470_00254 PE=4 SV=1
MM1 pKa = 7.91KK2 pKa = 10.17LRR4 pKa = 11.84NIISAVAAAAVAASAASLCAFADD27 pKa = 4.04DD28 pKa = 4.03QPAGYY33 pKa = 10.26VYY35 pKa = 10.35FMAEE39 pKa = 4.11KK40 pKa = 7.79TTLGQGFEE48 pKa = 4.18VEE50 pKa = 4.45PVKK53 pKa = 11.01VPYY56 pKa = 10.68YY57 pKa = 10.31EE58 pKa = 5.06GEE60 pKa = 4.01TGLDD64 pKa = 2.95IVEE67 pKa = 4.22RR68 pKa = 11.84AAEE71 pKa = 4.33IKK73 pKa = 10.02TEE75 pKa = 4.03DD76 pKa = 3.13AGYY79 pKa = 10.35GAFITAFADD88 pKa = 3.39TDD90 pKa = 4.28SNDD93 pKa = 3.16VVLPEE98 pKa = 4.38AVAEE102 pKa = 4.32VCTIASGRR110 pKa = 11.84TEE112 pKa = 4.32EE113 pKa = 4.58GWLSAYY119 pKa = 10.45DD120 pKa = 3.73YY121 pKa = 9.48TAEE124 pKa = 4.16SGWTYY129 pKa = 10.36FVNDD133 pKa = 3.96EE134 pKa = 4.12YY135 pKa = 11.47AQVGIADD142 pKa = 3.98YY143 pKa = 10.03TPADD147 pKa = 3.45GDD149 pKa = 4.05VVVFSFTVYY158 pKa = 10.72GYY160 pKa = 10.9GADD163 pKa = 3.52LGVDD167 pKa = 3.29NSSWGGAAALKK178 pKa = 9.78EE179 pKa = 4.05QVKK182 pKa = 8.12TAEE185 pKa = 4.21LVKK188 pKa = 10.8LFADD192 pKa = 4.84NKK194 pKa = 10.69DD195 pKa = 3.95LLNGSDD201 pKa = 3.52DD202 pKa = 3.31QAYY205 pKa = 10.24AFTAAGEE212 pKa = 4.02ALAQYY217 pKa = 10.21EE218 pKa = 4.57STQEE222 pKa = 4.08DD223 pKa = 3.3VDD225 pKa = 3.95NAVDD229 pKa = 3.51RR230 pKa = 11.84LEE232 pKa = 5.11NILGTEE238 pKa = 4.11TLVTEE243 pKa = 4.35EE244 pKa = 4.22XXXNSCDD251 pKa = 3.25RR252 pKa = 11.84GKK254 pKa = 9.96PLSS257 pKa = 3.67
MM1 pKa = 7.91KK2 pKa = 10.17LRR4 pKa = 11.84NIISAVAAAAVAASAASLCAFADD27 pKa = 4.04DD28 pKa = 4.03QPAGYY33 pKa = 10.26VYY35 pKa = 10.35FMAEE39 pKa = 4.11KK40 pKa = 7.79TTLGQGFEE48 pKa = 4.18VEE50 pKa = 4.45PVKK53 pKa = 11.01VPYY56 pKa = 10.68YY57 pKa = 10.31EE58 pKa = 5.06GEE60 pKa = 4.01TGLDD64 pKa = 2.95IVEE67 pKa = 4.22RR68 pKa = 11.84AAEE71 pKa = 4.33IKK73 pKa = 10.02TEE75 pKa = 4.03DD76 pKa = 3.13AGYY79 pKa = 10.35GAFITAFADD88 pKa = 3.39TDD90 pKa = 4.28SNDD93 pKa = 3.16VVLPEE98 pKa = 4.38AVAEE102 pKa = 4.32VCTIASGRR110 pKa = 11.84TEE112 pKa = 4.32EE113 pKa = 4.58GWLSAYY119 pKa = 10.45DD120 pKa = 3.73YY121 pKa = 9.48TAEE124 pKa = 4.16SGWTYY129 pKa = 10.36FVNDD133 pKa = 3.96EE134 pKa = 4.12YY135 pKa = 11.47AQVGIADD142 pKa = 3.98YY143 pKa = 10.03TPADD147 pKa = 3.45GDD149 pKa = 4.05VVVFSFTVYY158 pKa = 10.72GYY160 pKa = 10.9GADD163 pKa = 3.52LGVDD167 pKa = 3.29NSSWGGAAALKK178 pKa = 9.78EE179 pKa = 4.05QVKK182 pKa = 8.12TAEE185 pKa = 4.21LVKK188 pKa = 10.8LFADD192 pKa = 4.84NKK194 pKa = 10.69DD195 pKa = 3.95LLNGSDD201 pKa = 3.52DD202 pKa = 3.31QAYY205 pKa = 10.24AFTAAGEE212 pKa = 4.02ALAQYY217 pKa = 10.21EE218 pKa = 4.57STQEE222 pKa = 4.08DD223 pKa = 3.3VDD225 pKa = 3.95NAVDD229 pKa = 3.51RR230 pKa = 11.84LEE232 pKa = 5.11NILGTEE238 pKa = 4.11TLVTEE243 pKa = 4.35EE244 pKa = 4.22XXXNSCDD251 pKa = 3.25RR252 pKa = 11.84GKK254 pKa = 9.96PLSS257 pKa = 3.67
Molecular weight: 27.04 kDa
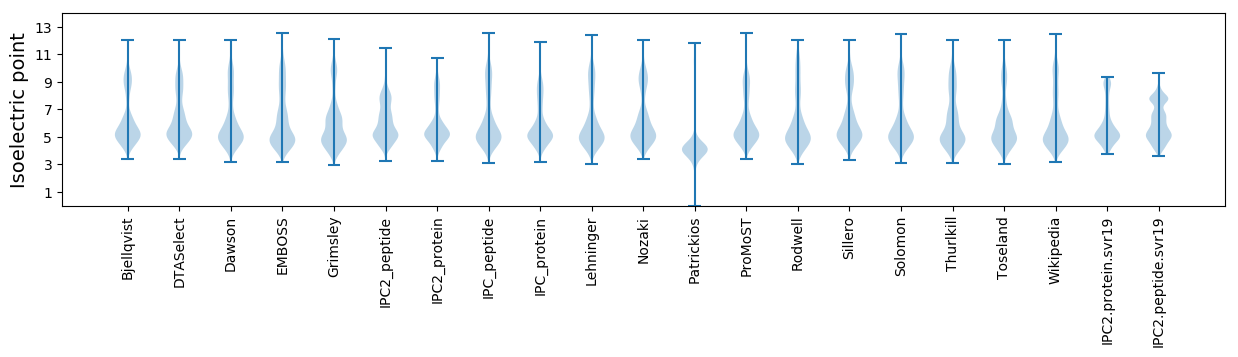
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5L4Q6|R5L4Q6_9FIRM Stage 0 sporulation protein A homolog OS=Eubacterium sp. CAG:115 OX=1262878 GN=BN470_00758 PE=4 SV=1
MM1 pKa = 7.2YY2 pKa = 10.32FLLRR6 pKa = 11.84RR7 pKa = 11.84IFLRR11 pKa = 11.84VDD13 pKa = 3.94LARR16 pKa = 11.84DD17 pKa = 3.47TVRR20 pKa = 11.84VSKK23 pKa = 10.86GLLLRR28 pKa = 11.84RR29 pKa = 11.84QFCIPFSAVTKK40 pKa = 9.63VEE42 pKa = 3.88VRR44 pKa = 11.84RR45 pKa = 11.84TPVLRR50 pKa = 11.84LLRR53 pKa = 11.84GKK55 pKa = 9.68RR56 pKa = 11.84VKK58 pKa = 10.4ISTLSGGVSFYY69 pKa = 10.61MKK71 pKa = 10.24AVEE74 pKa = 5.21HH75 pKa = 6.86IPFLPEE81 pKa = 3.2NDD83 pKa = 3.55GAYY86 pKa = 9.52IRR88 pKa = 11.84AKK90 pKa = 9.86PLQSVVGAFVDD101 pKa = 4.22TRR103 pKa = 11.84ALSGVITFGLFLNRR117 pKa = 11.84IGSVFGDD124 pKa = 3.51EE125 pKa = 4.21SFGRR129 pKa = 11.84IMTALAGAAEE139 pKa = 4.7GVTKK143 pKa = 10.79LLAALHH149 pKa = 6.2IGVPRR154 pKa = 11.84VTAFAAVFVGVAWLFVFLGKK174 pKa = 10.47ALGMLRR180 pKa = 11.84FRR182 pKa = 11.84LSCEE186 pKa = 3.8SGFITVQRR194 pKa = 11.84GILTLYY200 pKa = 9.48EE201 pKa = 4.1CRR203 pKa = 11.84LVRR206 pKa = 11.84NNLTGVLRR214 pKa = 11.84CDD216 pKa = 3.44TLTTLLLRR224 pKa = 11.84AAPLYY229 pKa = 10.9AHH231 pKa = 7.36DD232 pKa = 4.57VLLFPPVSRR241 pKa = 11.84RR242 pKa = 11.84TAEE245 pKa = 4.27RR246 pKa = 11.84ILTKK250 pKa = 9.52MCRR253 pKa = 11.84IPPPSGHH260 pKa = 5.95VKK262 pKa = 10.27PPFSAMFGHH271 pKa = 7.15CAAPLGWLGAFTAALLLTFIAEE293 pKa = 4.2PFAAGLVQSVLWCGIVVSLWFTAAYY318 pKa = 9.05AVYY321 pKa = 9.45MRR323 pKa = 11.84LSWLSSCPVEE333 pKa = 3.87TAGISFRR340 pKa = 11.84RR341 pKa = 11.84GARR344 pKa = 11.84LYY346 pKa = 8.84TAVMPAQKK354 pKa = 10.32LAFTITGRR362 pKa = 11.84NIFQRR367 pKa = 11.84FSGMCDD373 pKa = 2.49ITLCSAGRR381 pKa = 11.84KK382 pKa = 7.19QYY384 pKa = 11.05RR385 pKa = 11.84LRR387 pKa = 11.84NVPYY391 pKa = 10.6DD392 pKa = 3.37EE393 pKa = 4.31VRR395 pKa = 11.84RR396 pKa = 11.84IFPPEE401 pKa = 3.68KK402 pKa = 9.88PAA404 pKa = 4.96
MM1 pKa = 7.2YY2 pKa = 10.32FLLRR6 pKa = 11.84RR7 pKa = 11.84IFLRR11 pKa = 11.84VDD13 pKa = 3.94LARR16 pKa = 11.84DD17 pKa = 3.47TVRR20 pKa = 11.84VSKK23 pKa = 10.86GLLLRR28 pKa = 11.84RR29 pKa = 11.84QFCIPFSAVTKK40 pKa = 9.63VEE42 pKa = 3.88VRR44 pKa = 11.84RR45 pKa = 11.84TPVLRR50 pKa = 11.84LLRR53 pKa = 11.84GKK55 pKa = 9.68RR56 pKa = 11.84VKK58 pKa = 10.4ISTLSGGVSFYY69 pKa = 10.61MKK71 pKa = 10.24AVEE74 pKa = 5.21HH75 pKa = 6.86IPFLPEE81 pKa = 3.2NDD83 pKa = 3.55GAYY86 pKa = 9.52IRR88 pKa = 11.84AKK90 pKa = 9.86PLQSVVGAFVDD101 pKa = 4.22TRR103 pKa = 11.84ALSGVITFGLFLNRR117 pKa = 11.84IGSVFGDD124 pKa = 3.51EE125 pKa = 4.21SFGRR129 pKa = 11.84IMTALAGAAEE139 pKa = 4.7GVTKK143 pKa = 10.79LLAALHH149 pKa = 6.2IGVPRR154 pKa = 11.84VTAFAAVFVGVAWLFVFLGKK174 pKa = 10.47ALGMLRR180 pKa = 11.84FRR182 pKa = 11.84LSCEE186 pKa = 3.8SGFITVQRR194 pKa = 11.84GILTLYY200 pKa = 9.48EE201 pKa = 4.1CRR203 pKa = 11.84LVRR206 pKa = 11.84NNLTGVLRR214 pKa = 11.84CDD216 pKa = 3.44TLTTLLLRR224 pKa = 11.84AAPLYY229 pKa = 10.9AHH231 pKa = 7.36DD232 pKa = 4.57VLLFPPVSRR241 pKa = 11.84RR242 pKa = 11.84TAEE245 pKa = 4.27RR246 pKa = 11.84ILTKK250 pKa = 9.52MCRR253 pKa = 11.84IPPPSGHH260 pKa = 5.95VKK262 pKa = 10.27PPFSAMFGHH271 pKa = 7.15CAAPLGWLGAFTAALLLTFIAEE293 pKa = 4.2PFAAGLVQSVLWCGIVVSLWFTAAYY318 pKa = 9.05AVYY321 pKa = 9.45MRR323 pKa = 11.84LSWLSSCPVEE333 pKa = 3.87TAGISFRR340 pKa = 11.84RR341 pKa = 11.84GARR344 pKa = 11.84LYY346 pKa = 8.84TAVMPAQKK354 pKa = 10.32LAFTITGRR362 pKa = 11.84NIFQRR367 pKa = 11.84FSGMCDD373 pKa = 2.49ITLCSAGRR381 pKa = 11.84KK382 pKa = 7.19QYY384 pKa = 11.05RR385 pKa = 11.84LRR387 pKa = 11.84NVPYY391 pKa = 10.6DD392 pKa = 3.37EE393 pKa = 4.31VRR395 pKa = 11.84RR396 pKa = 11.84IFPPEE401 pKa = 3.68KK402 pKa = 9.88PAA404 pKa = 4.96
Molecular weight: 44.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
716508 |
31 |
2198 |
313.0 |
34.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.807 ± 0.067 | 1.709 ± 0.023 |
5.925 ± 0.045 | 6.95 ± 0.051 |
4.15 ± 0.035 | 7.568 ± 0.05 |
1.579 ± 0.024 | 6.875 ± 0.044 |
5.796 ± 0.05 | 8.567 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.905 ± 0.027 | 4.167 ± 0.04 |
3.665 ± 0.033 | 2.8 ± 0.029 |
4.829 ± 0.045 | 6.587 ± 0.047 |
5.581 ± 0.039 | 7.007 ± 0.04 |
0.838 ± 0.017 | 3.691 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
