
Lake Sarah-associated circular virus-35
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
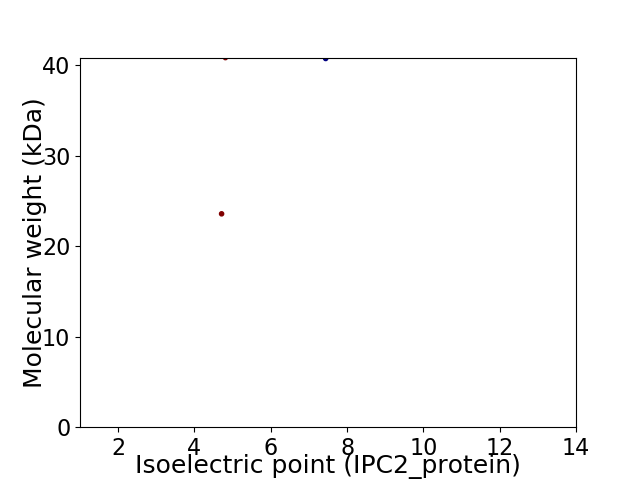
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQQ3|A0A140AQQ3_9VIRU Coat protein OS=Lake Sarah-associated circular virus-35 OX=1685763 PE=4 SV=1
MM1 pKa = 7.28YY2 pKa = 9.91APPEE6 pKa = 3.98ISTIQPPQAEE16 pKa = 4.24LCYY19 pKa = 10.06YY20 pKa = 10.45TMLSQLPGMKK30 pKa = 9.64TYY32 pKa = 10.79DD33 pKa = 3.55ILFGDD38 pKa = 5.02LDD40 pKa = 3.92TANFFPLLDD49 pKa = 5.43GITQGTGPHH58 pKa = 5.13QRR60 pKa = 11.84VGTSILLHH68 pKa = 6.15GIEE71 pKa = 4.14MRR73 pKa = 11.84FFTKK77 pKa = 10.61LQDD80 pKa = 3.57NEE82 pKa = 3.92QDD84 pKa = 3.66VVSFNARR91 pKa = 11.84FILVRR96 pKa = 11.84SLAGSKK102 pKa = 10.0PLRR105 pKa = 11.84DD106 pKa = 3.95DD107 pKa = 3.44VLEE110 pKa = 4.31GLVSSMIAHH119 pKa = 6.04YY120 pKa = 10.92NSDD123 pKa = 4.23FVPDD127 pKa = 4.48LYY129 pKa = 11.33EE130 pKa = 3.99PFYY133 pKa = 11.25DD134 pKa = 3.85EE135 pKa = 5.76YY136 pKa = 10.74IPEE139 pKa = 4.39MYY141 pKa = 9.76VQNPVLPVQSYY152 pKa = 9.35RR153 pKa = 11.84NFYY156 pKa = 10.75RR157 pKa = 11.84SFPVSWRR164 pKa = 11.84VVYY167 pKa = 10.0PRR169 pKa = 11.84DD170 pKa = 3.5NMDD173 pKa = 3.46PQKK176 pKa = 10.72IAGNVYY182 pKa = 10.57ALNLSDD188 pKa = 4.74FSSSIPAINVRR199 pKa = 11.84FYY201 pKa = 10.53YY202 pKa = 9.98TDD204 pKa = 3.15RR205 pKa = 5.91
MM1 pKa = 7.28YY2 pKa = 9.91APPEE6 pKa = 3.98ISTIQPPQAEE16 pKa = 4.24LCYY19 pKa = 10.06YY20 pKa = 10.45TMLSQLPGMKK30 pKa = 9.64TYY32 pKa = 10.79DD33 pKa = 3.55ILFGDD38 pKa = 5.02LDD40 pKa = 3.92TANFFPLLDD49 pKa = 5.43GITQGTGPHH58 pKa = 5.13QRR60 pKa = 11.84VGTSILLHH68 pKa = 6.15GIEE71 pKa = 4.14MRR73 pKa = 11.84FFTKK77 pKa = 10.61LQDD80 pKa = 3.57NEE82 pKa = 3.92QDD84 pKa = 3.66VVSFNARR91 pKa = 11.84FILVRR96 pKa = 11.84SLAGSKK102 pKa = 10.0PLRR105 pKa = 11.84DD106 pKa = 3.95DD107 pKa = 3.44VLEE110 pKa = 4.31GLVSSMIAHH119 pKa = 6.04YY120 pKa = 10.92NSDD123 pKa = 4.23FVPDD127 pKa = 4.48LYY129 pKa = 11.33EE130 pKa = 3.99PFYY133 pKa = 11.25DD134 pKa = 3.85EE135 pKa = 5.76YY136 pKa = 10.74IPEE139 pKa = 4.39MYY141 pKa = 9.76VQNPVLPVQSYY152 pKa = 9.35RR153 pKa = 11.84NFYY156 pKa = 10.75RR157 pKa = 11.84SFPVSWRR164 pKa = 11.84VVYY167 pKa = 10.0PRR169 pKa = 11.84DD170 pKa = 3.5NMDD173 pKa = 3.46PQKK176 pKa = 10.72IAGNVYY182 pKa = 10.57ALNLSDD188 pKa = 4.74FSSSIPAINVRR199 pKa = 11.84FYY201 pKa = 10.53YY202 pKa = 9.98TDD204 pKa = 3.15RR205 pKa = 5.91
Molecular weight: 23.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQQ3|A0A140AQQ3_9VIRU Coat protein OS=Lake Sarah-associated circular virus-35 OX=1685763 PE=4 SV=1
MM1 pKa = 6.96YY2 pKa = 10.19GVRR5 pKa = 11.84KK6 pKa = 9.23EE7 pKa = 4.19VFRR10 pKa = 11.84SAMTTAPARR19 pKa = 11.84RR20 pKa = 11.84PNSTRR25 pKa = 11.84TRR27 pKa = 11.84TRR29 pKa = 11.84VGQRR33 pKa = 11.84LRR35 pKa = 11.84RR36 pKa = 11.84FCFTINNYY44 pKa = 8.63TEE46 pKa = 4.0ALYY49 pKa = 11.03DD50 pKa = 3.74EE51 pKa = 5.19LLAFAVTTSWFVVGKK66 pKa = 9.93EE67 pKa = 3.78VGEE70 pKa = 4.25NGTPHH75 pKa = 6.6LQGACILNNQLSFNSLKK92 pKa = 11.0KK93 pKa = 9.06MDD95 pKa = 4.37CFARR99 pKa = 11.84AHH101 pKa = 6.26IEE103 pKa = 4.11TMKK106 pKa = 10.88GSPQDD111 pKa = 3.47NLVYY115 pKa = 9.52CTKK118 pKa = 10.32EE119 pKa = 3.61DD120 pKa = 3.57TNAYY124 pKa = 10.23VYY126 pKa = 8.75GTLPEE131 pKa = 4.58PGKK134 pKa = 11.09RR135 pKa = 11.84NDD137 pKa = 3.08LHH139 pKa = 6.38EE140 pKa = 4.57AVEE143 pKa = 4.35KK144 pKa = 10.6VKK146 pKa = 10.95EE147 pKa = 4.12GATLRR152 pKa = 11.84DD153 pKa = 3.5LALGDD158 pKa = 3.79GGPAVVKK165 pKa = 8.13FHH167 pKa = 7.24KK168 pKa = 10.49GLTVLRR174 pKa = 11.84SLCSTPRR181 pKa = 11.84ADD183 pKa = 3.68PPTVVWIYY191 pKa = 11.17GPTGVHH197 pKa = 5.49KK198 pKa = 9.05TRR200 pKa = 11.84CALEE204 pKa = 3.76FGKK207 pKa = 10.48ILGAASGSSEE217 pKa = 4.25ANSVWISGGGFRR229 pKa = 11.84WFDD232 pKa = 3.68GYY234 pKa = 11.15DD235 pKa = 3.63GQPVAIFDD243 pKa = 4.08DD244 pKa = 5.09FRR246 pKa = 11.84AKK248 pKa = 10.36QLTGAGGFTFLLRR261 pKa = 11.84LLDD264 pKa = 4.36RR265 pKa = 11.84YY266 pKa = 9.72PISVEE271 pKa = 3.99FKK273 pKa = 10.27GGFVNWTPRR282 pKa = 11.84IIFITTPHH290 pKa = 6.69DD291 pKa = 3.56VATTFAKK298 pKa = 10.35RR299 pKa = 11.84SQHH302 pKa = 5.57IPEE305 pKa = 5.93DD306 pKa = 3.24IAQLDD311 pKa = 3.5RR312 pKa = 11.84RR313 pKa = 11.84INKK316 pKa = 9.07VIHH319 pKa = 5.91FPEE322 pKa = 4.62VPSDD326 pKa = 3.65AVKK329 pKa = 11.1LEE331 pKa = 4.16LLDD334 pKa = 5.49ALTLCAGLRR343 pKa = 11.84PAGPQVPATIPGWNEE358 pKa = 3.52DD359 pKa = 3.67VEE361 pKa = 4.69LSGTASDD368 pKa = 5.04GEE370 pKa = 4.32
MM1 pKa = 6.96YY2 pKa = 10.19GVRR5 pKa = 11.84KK6 pKa = 9.23EE7 pKa = 4.19VFRR10 pKa = 11.84SAMTTAPARR19 pKa = 11.84RR20 pKa = 11.84PNSTRR25 pKa = 11.84TRR27 pKa = 11.84TRR29 pKa = 11.84VGQRR33 pKa = 11.84LRR35 pKa = 11.84RR36 pKa = 11.84FCFTINNYY44 pKa = 8.63TEE46 pKa = 4.0ALYY49 pKa = 11.03DD50 pKa = 3.74EE51 pKa = 5.19LLAFAVTTSWFVVGKK66 pKa = 9.93EE67 pKa = 3.78VGEE70 pKa = 4.25NGTPHH75 pKa = 6.6LQGACILNNQLSFNSLKK92 pKa = 11.0KK93 pKa = 9.06MDD95 pKa = 4.37CFARR99 pKa = 11.84AHH101 pKa = 6.26IEE103 pKa = 4.11TMKK106 pKa = 10.88GSPQDD111 pKa = 3.47NLVYY115 pKa = 9.52CTKK118 pKa = 10.32EE119 pKa = 3.61DD120 pKa = 3.57TNAYY124 pKa = 10.23VYY126 pKa = 8.75GTLPEE131 pKa = 4.58PGKK134 pKa = 11.09RR135 pKa = 11.84NDD137 pKa = 3.08LHH139 pKa = 6.38EE140 pKa = 4.57AVEE143 pKa = 4.35KK144 pKa = 10.6VKK146 pKa = 10.95EE147 pKa = 4.12GATLRR152 pKa = 11.84DD153 pKa = 3.5LALGDD158 pKa = 3.79GGPAVVKK165 pKa = 8.13FHH167 pKa = 7.24KK168 pKa = 10.49GLTVLRR174 pKa = 11.84SLCSTPRR181 pKa = 11.84ADD183 pKa = 3.68PPTVVWIYY191 pKa = 11.17GPTGVHH197 pKa = 5.49KK198 pKa = 9.05TRR200 pKa = 11.84CALEE204 pKa = 3.76FGKK207 pKa = 10.48ILGAASGSSEE217 pKa = 4.25ANSVWISGGGFRR229 pKa = 11.84WFDD232 pKa = 3.68GYY234 pKa = 11.15DD235 pKa = 3.63GQPVAIFDD243 pKa = 4.08DD244 pKa = 5.09FRR246 pKa = 11.84AKK248 pKa = 10.36QLTGAGGFTFLLRR261 pKa = 11.84LLDD264 pKa = 4.36RR265 pKa = 11.84YY266 pKa = 9.72PISVEE271 pKa = 3.99FKK273 pKa = 10.27GGFVNWTPRR282 pKa = 11.84IIFITTPHH290 pKa = 6.69DD291 pKa = 3.56VATTFAKK298 pKa = 10.35RR299 pKa = 11.84SQHH302 pKa = 5.57IPEE305 pKa = 5.93DD306 pKa = 3.24IAQLDD311 pKa = 3.5RR312 pKa = 11.84RR313 pKa = 11.84INKK316 pKa = 9.07VIHH319 pKa = 5.91FPEE322 pKa = 4.62VPSDD326 pKa = 3.65AVKK329 pKa = 11.1LEE331 pKa = 4.16LLDD334 pKa = 5.49ALTLCAGLRR343 pKa = 11.84PAGPQVPATIPGWNEE358 pKa = 3.52DD359 pKa = 3.67VEE361 pKa = 4.69LSGTASDD368 pKa = 5.04GEE370 pKa = 4.32
Molecular weight: 40.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
575 |
205 |
370 |
287.5 |
32.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.957 ± 1.519 | 1.391 ± 0.535 |
6.087 ± 0.728 | 4.87 ± 0.572 |
5.739 ± 0.357 | 7.652 ± 1.642 |
1.913 ± 0.266 | 4.87 ± 0.582 |
3.826 ± 1.11 | 8.87 ± 0.236 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.913 ± 0.889 | 4.174 ± 0.417 |
6.783 ± 0.894 | 3.304 ± 0.931 |
6.261 ± 0.53 | 5.913 ± 1.12 |
6.783 ± 1.416 | 7.304 ± 0.008 |
1.217 ± 0.432 | 4.174 ± 1.86 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
