
Halanaerobium salsuginis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Halanaerobiales; Halanaerobiaceae; Halanaerobium
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
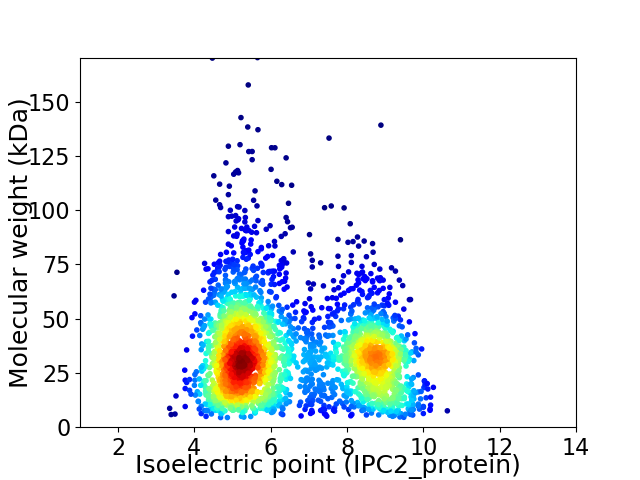
Virtual 2D-PAGE plot for 2783 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4FPU0|A0A1I4FPU0_9FIRM D-beta-D-heptose 1-phosphate adenylyltransferase OS=Halanaerobium salsuginis OX=29563 GN=SAMN02983006_00432 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.49KK3 pKa = 9.03LTITIALLLVVAFAMPVFGASFSDD27 pKa = 3.87VPSDD31 pKa = 2.76HH32 pKa = 6.84WAYY35 pKa = 11.05SAINKK40 pKa = 8.93LVAAGIVEE48 pKa = 4.84GYY50 pKa = 9.77PDD52 pKa = 4.63GEE54 pKa = 4.4FKK56 pKa = 10.87GQQSMTRR63 pKa = 11.84YY64 pKa = 8.92EE65 pKa = 4.09MAVMVSRR72 pKa = 11.84ALDD75 pKa = 3.99NIADD79 pKa = 4.09EE80 pKa = 4.55QKK82 pKa = 11.09EE83 pKa = 4.11MAAGLTTGQAEE94 pKa = 4.49DD95 pKa = 3.19VTAIVKK101 pKa = 10.51ALMEE105 pKa = 4.83KK106 pKa = 8.95NTSEE110 pKa = 4.34EE111 pKa = 4.0FTDD114 pKa = 3.78AQAKK118 pKa = 9.2KK119 pKa = 10.48VADD122 pKa = 3.41IVDD125 pKa = 3.58ALTFEE130 pKa = 5.04LKK132 pKa = 10.91AEE134 pKa = 4.24LKK136 pKa = 10.72VLGAKK141 pKa = 9.81VDD143 pKa = 3.89TLGKK147 pKa = 10.43DD148 pKa = 3.06VDD150 pKa = 4.51EE151 pKa = 5.07IKK153 pKa = 10.44ATIAALDD160 pKa = 3.74VPEE163 pKa = 5.13DD164 pKa = 3.84NIEE167 pKa = 3.94FSGVVTTYY175 pKa = 11.11AQVGSYY181 pKa = 11.24GDD183 pKa = 4.73DD184 pKa = 3.64DD185 pKa = 4.2EE186 pKa = 4.73QAATAEE192 pKa = 4.14ALADD196 pKa = 4.09GDD198 pKa = 4.64VLDD201 pKa = 6.01ADD203 pKa = 4.76LSNNGGGDD211 pKa = 3.61YY212 pKa = 11.03DD213 pKa = 5.36DD214 pKa = 6.22FPAEE218 pKa = 3.52QAFYY222 pKa = 11.2QEE224 pKa = 4.83IDD226 pKa = 3.38LLLNGSLKK234 pKa = 10.89GINFDD239 pKa = 4.61LAVDD243 pKa = 4.62TITNGFTDD251 pKa = 4.11YY252 pKa = 11.25EE253 pKa = 4.7GYY255 pKa = 11.01NYY257 pKa = 10.74DD258 pKa = 5.23DD259 pKa = 3.63YY260 pKa = 11.77TKK262 pKa = 11.05VLGEE266 pKa = 4.01QGDD269 pKa = 4.09EE270 pKa = 4.17NGIDD274 pKa = 3.73MDD276 pKa = 4.47TALLTITQDD285 pKa = 2.46NWTVKK290 pKa = 10.09IGDD293 pKa = 3.68MGDD296 pKa = 3.44YY297 pKa = 10.98DD298 pKa = 4.67VDD300 pKa = 3.92DD301 pKa = 3.56SAYY304 pKa = 10.51FYY306 pKa = 10.96DD307 pKa = 4.13VEE309 pKa = 4.26DD310 pKa = 4.15AEE312 pKa = 5.39GIEE315 pKa = 4.51FTGPVYY321 pKa = 10.96GFGVKK326 pKa = 10.09AFAVGYY332 pKa = 10.55DD333 pKa = 4.03DD334 pKa = 5.92GNLDD338 pKa = 4.04EE339 pKa = 5.74FATSDD344 pKa = 3.36DD345 pKa = 4.0LYY347 pKa = 11.34YY348 pKa = 11.06GVAVSKK354 pKa = 10.86EE355 pKa = 3.85FMNNTVVTGKK365 pKa = 10.51VYY367 pKa = 10.58QADD370 pKa = 4.05LDD372 pKa = 4.15SADD375 pKa = 4.12EE376 pKa = 4.27KK377 pKa = 10.82VTDD380 pKa = 3.42IAVAVSTDD388 pKa = 3.45LTDD391 pKa = 3.77SLTLSGEE398 pKa = 3.99AVYY401 pKa = 11.23NEE403 pKa = 4.63LDD405 pKa = 3.67EE406 pKa = 6.74ADD408 pKa = 4.26TDD410 pKa = 4.01DD411 pKa = 5.74SMFKK415 pKa = 10.2IAANYY420 pKa = 9.85IYY422 pKa = 10.32TDD424 pKa = 3.52TLSFRR429 pKa = 11.84GSFQTVGDD437 pKa = 4.11EE438 pKa = 4.33FNGGVDD444 pKa = 3.56HH445 pKa = 7.0FADD448 pKa = 3.53VQADD452 pKa = 3.67MEE454 pKa = 4.41EE455 pKa = 5.48DD456 pKa = 3.7YY457 pKa = 11.72DD458 pKa = 4.13FDD460 pKa = 5.35KK461 pKa = 10.92YY462 pKa = 9.68TLGTTYY468 pKa = 10.73VVNTNNSLDD477 pKa = 3.63FDD479 pKa = 4.34VAMVSYY485 pKa = 10.83DD486 pKa = 3.36ADD488 pKa = 3.73TAFSQDD494 pKa = 3.14FVDD497 pKa = 4.65DD498 pKa = 4.85LGYY501 pKa = 11.06SSDD504 pKa = 3.72TDD506 pKa = 3.4EE507 pKa = 5.37DD508 pKa = 4.18KK509 pKa = 11.37YY510 pKa = 11.0IYY512 pKa = 10.45SIGWNNTYY520 pKa = 10.97GKK522 pKa = 8.64FTNHH526 pKa = 5.59VGLEE530 pKa = 4.19YY531 pKa = 11.09VEE533 pKa = 4.05NDD535 pKa = 3.48YY536 pKa = 10.07YY537 pKa = 11.18TDD539 pKa = 3.57EE540 pKa = 4.58TDD542 pKa = 4.94DD543 pKa = 3.6NTTVIEE549 pKa = 4.62LGTVYY554 pKa = 10.91DD555 pKa = 3.92WTAKK559 pKa = 9.29TDD561 pKa = 3.6LRR563 pKa = 11.84AKK565 pKa = 9.91IVNKK569 pKa = 10.36NADD572 pKa = 3.93LYY574 pKa = 11.36DD575 pKa = 4.44NSDD578 pKa = 3.25VWDD581 pKa = 3.48KK582 pKa = 11.96DD583 pKa = 3.33MNFTYY588 pKa = 10.36LQAGIDD594 pKa = 3.66HH595 pKa = 6.34QLADD599 pKa = 4.47NISWITDD606 pKa = 3.24VQYY609 pKa = 9.93ITGDD613 pKa = 3.04VDD615 pKa = 3.43QLTNHH620 pKa = 6.37SADD623 pKa = 4.07DD624 pKa = 4.0SVDD627 pKa = 3.6SYY629 pKa = 11.67VANGDD634 pKa = 3.59NADD637 pKa = 3.46INGNFITTQLSVSFF651 pKa = 4.46
MM1 pKa = 7.59KK2 pKa = 10.49KK3 pKa = 9.03LTITIALLLVVAFAMPVFGASFSDD27 pKa = 3.87VPSDD31 pKa = 2.76HH32 pKa = 6.84WAYY35 pKa = 11.05SAINKK40 pKa = 8.93LVAAGIVEE48 pKa = 4.84GYY50 pKa = 9.77PDD52 pKa = 4.63GEE54 pKa = 4.4FKK56 pKa = 10.87GQQSMTRR63 pKa = 11.84YY64 pKa = 8.92EE65 pKa = 4.09MAVMVSRR72 pKa = 11.84ALDD75 pKa = 3.99NIADD79 pKa = 4.09EE80 pKa = 4.55QKK82 pKa = 11.09EE83 pKa = 4.11MAAGLTTGQAEE94 pKa = 4.49DD95 pKa = 3.19VTAIVKK101 pKa = 10.51ALMEE105 pKa = 4.83KK106 pKa = 8.95NTSEE110 pKa = 4.34EE111 pKa = 4.0FTDD114 pKa = 3.78AQAKK118 pKa = 9.2KK119 pKa = 10.48VADD122 pKa = 3.41IVDD125 pKa = 3.58ALTFEE130 pKa = 5.04LKK132 pKa = 10.91AEE134 pKa = 4.24LKK136 pKa = 10.72VLGAKK141 pKa = 9.81VDD143 pKa = 3.89TLGKK147 pKa = 10.43DD148 pKa = 3.06VDD150 pKa = 4.51EE151 pKa = 5.07IKK153 pKa = 10.44ATIAALDD160 pKa = 3.74VPEE163 pKa = 5.13DD164 pKa = 3.84NIEE167 pKa = 3.94FSGVVTTYY175 pKa = 11.11AQVGSYY181 pKa = 11.24GDD183 pKa = 4.73DD184 pKa = 3.64DD185 pKa = 4.2EE186 pKa = 4.73QAATAEE192 pKa = 4.14ALADD196 pKa = 4.09GDD198 pKa = 4.64VLDD201 pKa = 6.01ADD203 pKa = 4.76LSNNGGGDD211 pKa = 3.61YY212 pKa = 11.03DD213 pKa = 5.36DD214 pKa = 6.22FPAEE218 pKa = 3.52QAFYY222 pKa = 11.2QEE224 pKa = 4.83IDD226 pKa = 3.38LLLNGSLKK234 pKa = 10.89GINFDD239 pKa = 4.61LAVDD243 pKa = 4.62TITNGFTDD251 pKa = 4.11YY252 pKa = 11.25EE253 pKa = 4.7GYY255 pKa = 11.01NYY257 pKa = 10.74DD258 pKa = 5.23DD259 pKa = 3.63YY260 pKa = 11.77TKK262 pKa = 11.05VLGEE266 pKa = 4.01QGDD269 pKa = 4.09EE270 pKa = 4.17NGIDD274 pKa = 3.73MDD276 pKa = 4.47TALLTITQDD285 pKa = 2.46NWTVKK290 pKa = 10.09IGDD293 pKa = 3.68MGDD296 pKa = 3.44YY297 pKa = 10.98DD298 pKa = 4.67VDD300 pKa = 3.92DD301 pKa = 3.56SAYY304 pKa = 10.51FYY306 pKa = 10.96DD307 pKa = 4.13VEE309 pKa = 4.26DD310 pKa = 4.15AEE312 pKa = 5.39GIEE315 pKa = 4.51FTGPVYY321 pKa = 10.96GFGVKK326 pKa = 10.09AFAVGYY332 pKa = 10.55DD333 pKa = 4.03DD334 pKa = 5.92GNLDD338 pKa = 4.04EE339 pKa = 5.74FATSDD344 pKa = 3.36DD345 pKa = 4.0LYY347 pKa = 11.34YY348 pKa = 11.06GVAVSKK354 pKa = 10.86EE355 pKa = 3.85FMNNTVVTGKK365 pKa = 10.51VYY367 pKa = 10.58QADD370 pKa = 4.05LDD372 pKa = 4.15SADD375 pKa = 4.12EE376 pKa = 4.27KK377 pKa = 10.82VTDD380 pKa = 3.42IAVAVSTDD388 pKa = 3.45LTDD391 pKa = 3.77SLTLSGEE398 pKa = 3.99AVYY401 pKa = 11.23NEE403 pKa = 4.63LDD405 pKa = 3.67EE406 pKa = 6.74ADD408 pKa = 4.26TDD410 pKa = 4.01DD411 pKa = 5.74SMFKK415 pKa = 10.2IAANYY420 pKa = 9.85IYY422 pKa = 10.32TDD424 pKa = 3.52TLSFRR429 pKa = 11.84GSFQTVGDD437 pKa = 4.11EE438 pKa = 4.33FNGGVDD444 pKa = 3.56HH445 pKa = 7.0FADD448 pKa = 3.53VQADD452 pKa = 3.67MEE454 pKa = 4.41EE455 pKa = 5.48DD456 pKa = 3.7YY457 pKa = 11.72DD458 pKa = 4.13FDD460 pKa = 5.35KK461 pKa = 10.92YY462 pKa = 9.68TLGTTYY468 pKa = 10.73VVNTNNSLDD477 pKa = 3.63FDD479 pKa = 4.34VAMVSYY485 pKa = 10.83DD486 pKa = 3.36ADD488 pKa = 3.73TAFSQDD494 pKa = 3.14FVDD497 pKa = 4.65DD498 pKa = 4.85LGYY501 pKa = 11.06SSDD504 pKa = 3.72TDD506 pKa = 3.4EE507 pKa = 5.37DD508 pKa = 4.18KK509 pKa = 11.37YY510 pKa = 11.0IYY512 pKa = 10.45SIGWNNTYY520 pKa = 10.97GKK522 pKa = 8.64FTNHH526 pKa = 5.59VGLEE530 pKa = 4.19YY531 pKa = 11.09VEE533 pKa = 4.05NDD535 pKa = 3.48YY536 pKa = 10.07YY537 pKa = 11.18TDD539 pKa = 3.57EE540 pKa = 4.58TDD542 pKa = 4.94DD543 pKa = 3.6NTTVIEE549 pKa = 4.62LGTVYY554 pKa = 10.91DD555 pKa = 3.92WTAKK559 pKa = 9.29TDD561 pKa = 3.6LRR563 pKa = 11.84AKK565 pKa = 9.91IVNKK569 pKa = 10.36NADD572 pKa = 3.93LYY574 pKa = 11.36DD575 pKa = 4.44NSDD578 pKa = 3.25VWDD581 pKa = 3.48KK582 pKa = 11.96DD583 pKa = 3.33MNFTYY588 pKa = 10.36LQAGIDD594 pKa = 3.66HH595 pKa = 6.34QLADD599 pKa = 4.47NISWITDD606 pKa = 3.24VQYY609 pKa = 9.93ITGDD613 pKa = 3.04VDD615 pKa = 3.43QLTNHH620 pKa = 6.37SADD623 pKa = 4.07DD624 pKa = 4.0SVDD627 pKa = 3.6SYY629 pKa = 11.67VANGDD634 pKa = 3.59NADD637 pKa = 3.46INGNFITTQLSVSFF651 pKa = 4.46
Molecular weight: 71.37 kDa
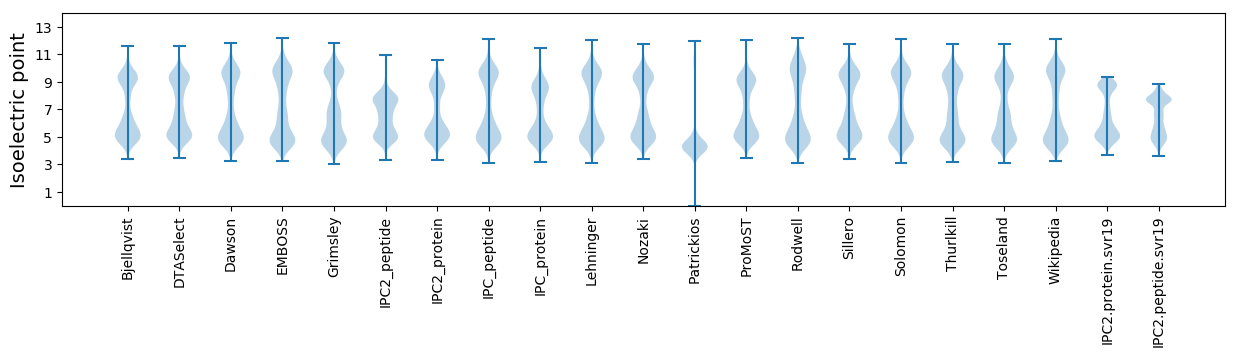
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4LIS9|A0A1I4LIS9_9FIRM Uncharacterized protein OS=Halanaerobium salsuginis OX=29563 GN=SAMN02983006_02333 PE=4 SV=1
MM1 pKa = 7.35ARR3 pKa = 11.84IEE5 pKa = 4.74GVDD8 pKa = 3.49LPRR11 pKa = 11.84NKK13 pKa = 9.85RR14 pKa = 11.84VEE16 pKa = 3.72IGLTYY21 pKa = 10.42IFGIGKK27 pKa = 8.41TSAQEE32 pKa = 3.61ILANTGVDD40 pKa = 3.31PDD42 pKa = 4.11TRR44 pKa = 11.84IKK46 pKa = 10.97DD47 pKa = 3.49LTEE50 pKa = 4.7AEE52 pKa = 4.18ISSLRR57 pKa = 11.84SEE59 pKa = 3.86IDD61 pKa = 2.69ANYY64 pKa = 9.6IVEE67 pKa = 4.35GEE69 pKa = 3.95LRR71 pKa = 11.84RR72 pKa = 11.84EE73 pKa = 3.44RR74 pKa = 11.84RR75 pKa = 11.84ADD77 pKa = 3.09IKK79 pKa = 10.57RR80 pKa = 11.84LKK82 pKa = 10.77DD83 pKa = 2.99IGCYY87 pKa = 9.37RR88 pKa = 11.84GLRR91 pKa = 11.84HH92 pKa = 6.72RR93 pKa = 11.84RR94 pKa = 11.84GLPVRR99 pKa = 11.84GQRR102 pKa = 11.84TKK104 pKa = 10.45TNARR108 pKa = 11.84TRR110 pKa = 11.84KK111 pKa = 8.76GPKK114 pKa = 8.14RR115 pKa = 11.84TVGVSRR121 pKa = 11.84VKK123 pKa = 10.8
MM1 pKa = 7.35ARR3 pKa = 11.84IEE5 pKa = 4.74GVDD8 pKa = 3.49LPRR11 pKa = 11.84NKK13 pKa = 9.85RR14 pKa = 11.84VEE16 pKa = 3.72IGLTYY21 pKa = 10.42IFGIGKK27 pKa = 8.41TSAQEE32 pKa = 3.61ILANTGVDD40 pKa = 3.31PDD42 pKa = 4.11TRR44 pKa = 11.84IKK46 pKa = 10.97DD47 pKa = 3.49LTEE50 pKa = 4.7AEE52 pKa = 4.18ISSLRR57 pKa = 11.84SEE59 pKa = 3.86IDD61 pKa = 2.69ANYY64 pKa = 9.6IVEE67 pKa = 4.35GEE69 pKa = 3.95LRR71 pKa = 11.84RR72 pKa = 11.84EE73 pKa = 3.44RR74 pKa = 11.84RR75 pKa = 11.84ADD77 pKa = 3.09IKK79 pKa = 10.57RR80 pKa = 11.84LKK82 pKa = 10.77DD83 pKa = 2.99IGCYY87 pKa = 9.37RR88 pKa = 11.84GLRR91 pKa = 11.84HH92 pKa = 6.72RR93 pKa = 11.84RR94 pKa = 11.84GLPVRR99 pKa = 11.84GQRR102 pKa = 11.84TKK104 pKa = 10.45TNARR108 pKa = 11.84TRR110 pKa = 11.84KK111 pKa = 8.76GPKK114 pKa = 8.14RR115 pKa = 11.84TVGVSRR121 pKa = 11.84VKK123 pKa = 10.8
Molecular weight: 13.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
888209 |
39 |
1537 |
319.2 |
35.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.847 ± 0.054 | 0.662 ± 0.013 |
5.304 ± 0.038 | 7.007 ± 0.052 |
4.535 ± 0.037 | 6.256 ± 0.04 |
1.467 ± 0.016 | 8.649 ± 0.047 |
7.771 ± 0.049 | 10.697 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.022 | 5.539 ± 0.045 |
3.209 ± 0.024 | 3.95 ± 0.034 |
3.652 ± 0.024 | 5.968 ± 0.032 |
4.698 ± 0.029 | 5.972 ± 0.035 |
0.817 ± 0.017 | 3.811 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
