
Clostridium clostridioforme CAG:132
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
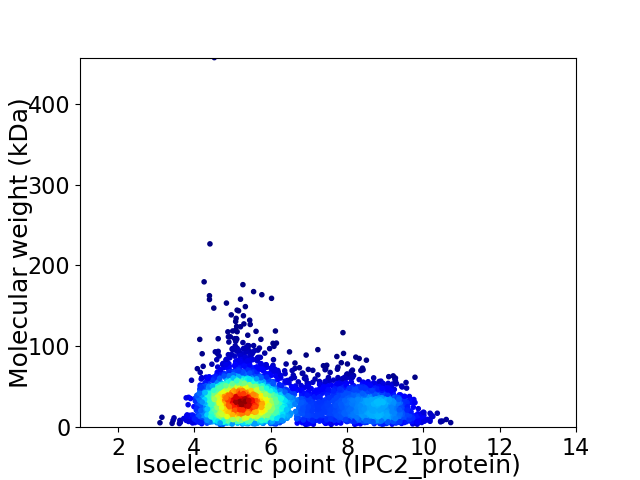
Virtual 2D-PAGE plot for 4152 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6JVG6|R6JVG6_9CLOT Aldedh domain-containing protein OS=Clostridium clostridioforme CAG:132 OX=1263065 GN=BN486_01988 PE=4 SV=1
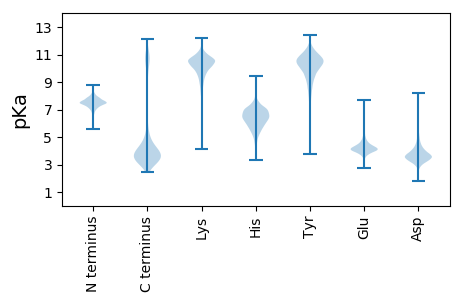
MM1 pKa = 7.6GRR3 pKa = 11.84LKK5 pKa = 10.13QQAVIWMSGALLMFMCGTAMASTRR29 pKa = 11.84TEE31 pKa = 3.68IDD33 pKa = 3.7SISLDD38 pKa = 3.35VEE40 pKa = 4.87SNIEE44 pKa = 4.19AGDD47 pKa = 3.6SSGDD51 pKa = 3.36VDD53 pKa = 4.13VTCDD57 pKa = 3.05SGDD60 pKa = 3.75YY61 pKa = 10.56YY62 pKa = 11.58VDD64 pKa = 5.08DD65 pKa = 4.34IEE67 pKa = 4.95ITNEE71 pKa = 4.15PKK73 pKa = 10.85NGWDD77 pKa = 4.91DD78 pKa = 3.54GDD80 pKa = 3.86KK81 pKa = 10.88PKK83 pKa = 11.22LKK85 pKa = 9.58VTVEE89 pKa = 4.17AEE91 pKa = 3.48DD92 pKa = 4.61DD93 pKa = 4.5YY94 pKa = 11.69YY95 pKa = 11.36FSSGLSKK102 pKa = 11.17NDD104 pKa = 2.94VDD106 pKa = 5.93LKK108 pKa = 10.98GADD111 pKa = 3.45GKK113 pKa = 9.33VTSVTRR119 pKa = 11.84KK120 pKa = 8.79SSTLIVYY127 pKa = 7.97ITLDD131 pKa = 3.62SLDD134 pKa = 4.56GSDD137 pKa = 4.35SGYY140 pKa = 11.24DD141 pKa = 3.29LDD143 pKa = 5.13VYY145 pKa = 10.83GLEE148 pKa = 4.15WDD150 pKa = 4.45EE151 pKa = 4.43SDD153 pKa = 6.1GMASWEE159 pKa = 4.14DD160 pKa = 3.18SGDD163 pKa = 3.27ARR165 pKa = 11.84KK166 pKa = 10.36YY167 pKa = 8.99EE168 pKa = 3.83VRR170 pKa = 11.84LYY172 pKa = 11.06RR173 pKa = 11.84NDD175 pKa = 3.48NSVTSVLTTSDD186 pKa = 2.78TSYY189 pKa = 11.36DD190 pKa = 3.33FSGYY194 pKa = 7.04ITKK197 pKa = 10.72SGDD200 pKa = 3.12YY201 pKa = 8.26MFKK204 pKa = 10.52VRR206 pKa = 11.84AVYY209 pKa = 10.42NSSDD213 pKa = 3.32KK214 pKa = 11.35GSWEE218 pKa = 4.49EE219 pKa = 3.69SDD221 pKa = 2.96SWYY224 pKa = 10.81VSSEE228 pKa = 3.99EE229 pKa = 4.19ADD231 pKa = 3.94EE232 pKa = 4.74ISDD235 pKa = 3.47GRR237 pKa = 11.84RR238 pKa = 11.84TYY240 pKa = 11.09GSSSSDD246 pKa = 3.32YY247 pKa = 10.61KK248 pKa = 11.17GAWLRR253 pKa = 11.84DD254 pKa = 3.56AVGWWYY260 pKa = 11.29CNADD264 pKa = 3.07KK265 pKa = 10.97SYY267 pKa = 9.96TVNNWQYY274 pKa = 11.27IDD276 pKa = 3.81DD277 pKa = 3.46RR278 pKa = 11.84WYY280 pKa = 9.85FFNAAGYY287 pKa = 8.04MVTGWIDD294 pKa = 3.02WNGRR298 pKa = 11.84WYY300 pKa = 11.2YY301 pKa = 11.01CGDD304 pKa = 4.28DD305 pKa = 3.07GAMYY309 pKa = 10.93YY310 pKa = 8.88DD311 pKa = 3.66TTTPDD316 pKa = 3.07GYY318 pKa = 11.53YY319 pKa = 10.71VGDD322 pKa = 4.24DD323 pKa = 4.06GAWVQQ328 pKa = 4.15
MM1 pKa = 7.6GRR3 pKa = 11.84LKK5 pKa = 10.13QQAVIWMSGALLMFMCGTAMASTRR29 pKa = 11.84TEE31 pKa = 3.68IDD33 pKa = 3.7SISLDD38 pKa = 3.35VEE40 pKa = 4.87SNIEE44 pKa = 4.19AGDD47 pKa = 3.6SSGDD51 pKa = 3.36VDD53 pKa = 4.13VTCDD57 pKa = 3.05SGDD60 pKa = 3.75YY61 pKa = 10.56YY62 pKa = 11.58VDD64 pKa = 5.08DD65 pKa = 4.34IEE67 pKa = 4.95ITNEE71 pKa = 4.15PKK73 pKa = 10.85NGWDD77 pKa = 4.91DD78 pKa = 3.54GDD80 pKa = 3.86KK81 pKa = 10.88PKK83 pKa = 11.22LKK85 pKa = 9.58VTVEE89 pKa = 4.17AEE91 pKa = 3.48DD92 pKa = 4.61DD93 pKa = 4.5YY94 pKa = 11.69YY95 pKa = 11.36FSSGLSKK102 pKa = 11.17NDD104 pKa = 2.94VDD106 pKa = 5.93LKK108 pKa = 10.98GADD111 pKa = 3.45GKK113 pKa = 9.33VTSVTRR119 pKa = 11.84KK120 pKa = 8.79SSTLIVYY127 pKa = 7.97ITLDD131 pKa = 3.62SLDD134 pKa = 4.56GSDD137 pKa = 4.35SGYY140 pKa = 11.24DD141 pKa = 3.29LDD143 pKa = 5.13VYY145 pKa = 10.83GLEE148 pKa = 4.15WDD150 pKa = 4.45EE151 pKa = 4.43SDD153 pKa = 6.1GMASWEE159 pKa = 4.14DD160 pKa = 3.18SGDD163 pKa = 3.27ARR165 pKa = 11.84KK166 pKa = 10.36YY167 pKa = 8.99EE168 pKa = 3.83VRR170 pKa = 11.84LYY172 pKa = 11.06RR173 pKa = 11.84NDD175 pKa = 3.48NSVTSVLTTSDD186 pKa = 2.78TSYY189 pKa = 11.36DD190 pKa = 3.33FSGYY194 pKa = 7.04ITKK197 pKa = 10.72SGDD200 pKa = 3.12YY201 pKa = 8.26MFKK204 pKa = 10.52VRR206 pKa = 11.84AVYY209 pKa = 10.42NSSDD213 pKa = 3.32KK214 pKa = 11.35GSWEE218 pKa = 4.49EE219 pKa = 3.69SDD221 pKa = 2.96SWYY224 pKa = 10.81VSSEE228 pKa = 3.99EE229 pKa = 4.19ADD231 pKa = 3.94EE232 pKa = 4.74ISDD235 pKa = 3.47GRR237 pKa = 11.84RR238 pKa = 11.84TYY240 pKa = 11.09GSSSSDD246 pKa = 3.32YY247 pKa = 10.61KK248 pKa = 11.17GAWLRR253 pKa = 11.84DD254 pKa = 3.56AVGWWYY260 pKa = 11.29CNADD264 pKa = 3.07KK265 pKa = 10.97SYY267 pKa = 9.96TVNNWQYY274 pKa = 11.27IDD276 pKa = 3.81DD277 pKa = 3.46RR278 pKa = 11.84WYY280 pKa = 9.85FFNAAGYY287 pKa = 8.04MVTGWIDD294 pKa = 3.02WNGRR298 pKa = 11.84WYY300 pKa = 11.2YY301 pKa = 11.01CGDD304 pKa = 4.28DD305 pKa = 3.07GAMYY309 pKa = 10.93YY310 pKa = 8.88DD311 pKa = 3.66TTTPDD316 pKa = 3.07GYY318 pKa = 11.53YY319 pKa = 10.71VGDD322 pKa = 4.24DD323 pKa = 4.06GAWVQQ328 pKa = 4.15
Molecular weight: 36.93 kDa
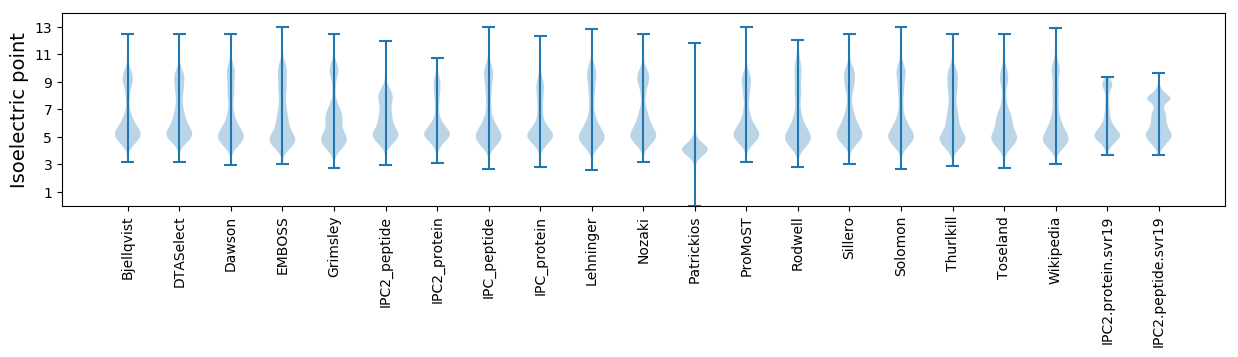
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6KFZ9|R6KFZ9_9CLOT SSD domain-containing protein OS=Clostridium clostridioforme CAG:132 OX=1263065 GN=BN486_01440 PE=4 SV=1
MM1 pKa = 7.13GVSRR5 pKa = 11.84KK6 pKa = 9.9SISVQSQMCGDD17 pKa = 3.74TQIVVLVIRR26 pKa = 11.84EE27 pKa = 3.95QNRR30 pKa = 11.84DD31 pKa = 3.07LKK33 pKa = 10.89RR34 pKa = 11.84AVARR38 pKa = 11.84TRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84LKK44 pKa = 10.35RR45 pKa = 11.84AALKK49 pKa = 10.3FGKK52 pKa = 9.7CLRR55 pKa = 11.84RR56 pKa = 11.84NWLYY60 pKa = 10.78ILIGLWLTWKK70 pKa = 10.62AIDD73 pKa = 3.35AAYY76 pKa = 10.04EE77 pKa = 3.91FRR79 pKa = 11.84GYY81 pKa = 10.31RR82 pKa = 11.84AIGGEE87 pKa = 4.09YY88 pKa = 10.24LVLPMFLALVVMIRR102 pKa = 11.84KK103 pKa = 6.23TVSFIEE109 pKa = 4.31YY110 pKa = 9.8CRR112 pKa = 11.84EE113 pKa = 3.81GG114 pKa = 3.35
MM1 pKa = 7.13GVSRR5 pKa = 11.84KK6 pKa = 9.9SISVQSQMCGDD17 pKa = 3.74TQIVVLVIRR26 pKa = 11.84EE27 pKa = 3.95QNRR30 pKa = 11.84DD31 pKa = 3.07LKK33 pKa = 10.89RR34 pKa = 11.84AVARR38 pKa = 11.84TRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84LKK44 pKa = 10.35RR45 pKa = 11.84AALKK49 pKa = 10.3FGKK52 pKa = 9.7CLRR55 pKa = 11.84RR56 pKa = 11.84NWLYY60 pKa = 10.78ILIGLWLTWKK70 pKa = 10.62AIDD73 pKa = 3.35AAYY76 pKa = 10.04EE77 pKa = 3.91FRR79 pKa = 11.84GYY81 pKa = 10.31RR82 pKa = 11.84AIGGEE87 pKa = 4.09YY88 pKa = 10.24LVLPMFLALVVMIRR102 pKa = 11.84KK103 pKa = 6.23TVSFIEE109 pKa = 4.31YY110 pKa = 9.8CRR112 pKa = 11.84EE113 pKa = 3.81GG114 pKa = 3.35
Molecular weight: 13.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1251376 |
29 |
4096 |
301.4 |
33.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.1 ± 0.049 | 1.588 ± 0.016 |
5.537 ± 0.03 | 7.073 ± 0.044 |
3.989 ± 0.027 | 8.015 ± 0.041 |
1.761 ± 0.017 | 6.864 ± 0.034 |
5.746 ± 0.029 | 9.08 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.481 ± 0.019 | 3.801 ± 0.026 |
3.657 ± 0.021 | 3.22 ± 0.023 |
5.068 ± 0.033 | 5.833 ± 0.028 |
5.199 ± 0.028 | 7.049 ± 0.033 |
0.991 ± 0.014 | 3.948 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
