
Ctenophore-associated circular genome 1
Taxonomy: Viruses; Satellites; DNA satellites; Single stranded DNA satellites
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
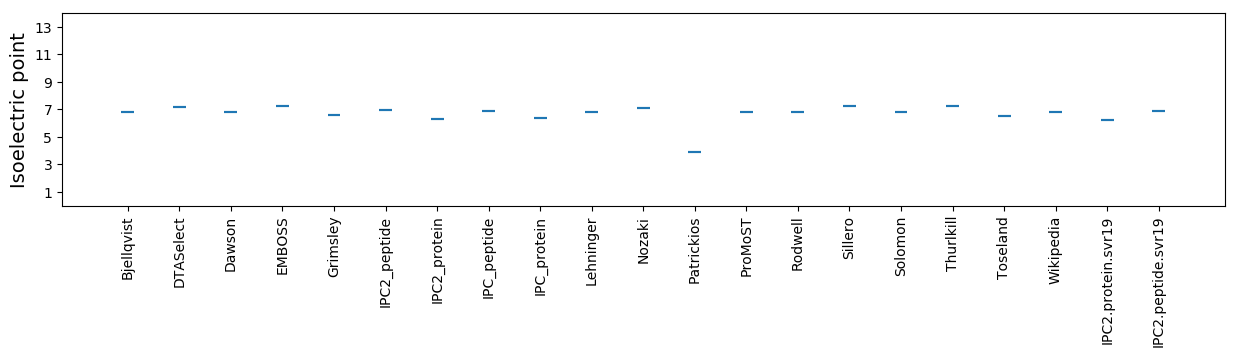
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A141MJB3|A0A141MJB3_9VIRU ATP-dependent helicase Rep OS=Ctenophore-associated circular genome 1 OX=1778570 PE=3 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.5LSDD6 pKa = 3.51NRR8 pKa = 11.84VVNYY12 pKa = 9.86VFTWNNYY19 pKa = 7.47TDD21 pKa = 3.82DD22 pKa = 3.67VKK24 pKa = 11.58AFIGNFANDD33 pKa = 3.29YY34 pKa = 9.95CKK36 pKa = 9.84WLVYY40 pKa = 10.54GEE42 pKa = 4.34EE43 pKa = 4.37VGASGTPHH51 pKa = 6.46LQGCFSLYY59 pKa = 9.92DD60 pKa = 3.67KK61 pKa = 11.2KK62 pKa = 10.99RR63 pKa = 11.84ITQLCKK69 pKa = 10.58LGFKK73 pKa = 10.45AHH75 pKa = 7.0LEE77 pKa = 3.99PMKK80 pKa = 10.66GSPKK84 pKa = 9.96QAIAYY89 pKa = 7.12CKK91 pKa = 9.95KK92 pKa = 10.41DD93 pKa = 3.27GSFTEE98 pKa = 4.64LGDD101 pKa = 3.47VTRR104 pKa = 11.84GQGEE108 pKa = 4.23RR109 pKa = 11.84NDD111 pKa = 3.73LKK113 pKa = 10.84RR114 pKa = 11.84ACEE117 pKa = 4.11LIKK120 pKa = 10.36EE121 pKa = 4.5GKK123 pKa = 9.75SISEE127 pKa = 4.25VADD130 pKa = 3.65ACPTTYY136 pKa = 10.67VKK138 pKa = 10.68YY139 pKa = 10.69SRR141 pKa = 11.84GLRR144 pKa = 11.84EE145 pKa = 3.85LALVASEE152 pKa = 4.61AYY154 pKa = 8.84EE155 pKa = 4.17HH156 pKa = 6.56EE157 pKa = 4.61SVRR160 pKa = 11.84GVWIYY165 pKa = 10.95GQTGVGKK172 pKa = 9.75SHH174 pKa = 7.96LARR177 pKa = 11.84DD178 pKa = 4.24VYY180 pKa = 11.26PNLFDD185 pKa = 4.77KK186 pKa = 11.08SQSKK190 pKa = 7.92WWDD193 pKa = 4.15GYY195 pKa = 10.7CNEE198 pKa = 4.43HH199 pKa = 6.15TVLIDD204 pKa = 6.14DD205 pKa = 5.98LDD207 pKa = 4.16TDD209 pKa = 4.6CLGHH213 pKa = 5.94YY214 pKa = 9.58LKK216 pKa = 10.65RR217 pKa = 11.84WADD220 pKa = 3.39KK221 pKa = 10.05YY222 pKa = 11.24ACTGEE227 pKa = 4.49TKK229 pKa = 10.72GGTIKK234 pKa = 10.46LQHH237 pKa = 6.25RR238 pKa = 11.84VFVVTSNYY246 pKa = 10.07LPWEE250 pKa = 3.94LHH252 pKa = 6.33KK253 pKa = 11.15DD254 pKa = 3.5DD255 pKa = 5.79LMAEE259 pKa = 4.68AISRR263 pKa = 11.84RR264 pKa = 11.84FTIYY268 pKa = 10.36HH269 pKa = 6.51KK270 pKa = 7.8EE271 pKa = 4.01TRR273 pKa = 11.84DD274 pKa = 3.47QKK276 pKa = 11.63LEE278 pKa = 4.1DD279 pKa = 4.42CEE281 pKa = 4.32VLTYY285 pKa = 10.93ADD287 pKa = 3.6RR288 pKa = 11.84HH289 pKa = 5.75NIII292 pKa = 4.56
MM1 pKa = 7.84PKK3 pKa = 10.5LSDD6 pKa = 3.51NRR8 pKa = 11.84VVNYY12 pKa = 9.86VFTWNNYY19 pKa = 7.47TDD21 pKa = 3.82DD22 pKa = 3.67VKK24 pKa = 11.58AFIGNFANDD33 pKa = 3.29YY34 pKa = 9.95CKK36 pKa = 9.84WLVYY40 pKa = 10.54GEE42 pKa = 4.34EE43 pKa = 4.37VGASGTPHH51 pKa = 6.46LQGCFSLYY59 pKa = 9.92DD60 pKa = 3.67KK61 pKa = 11.2KK62 pKa = 10.99RR63 pKa = 11.84ITQLCKK69 pKa = 10.58LGFKK73 pKa = 10.45AHH75 pKa = 7.0LEE77 pKa = 3.99PMKK80 pKa = 10.66GSPKK84 pKa = 9.96QAIAYY89 pKa = 7.12CKK91 pKa = 9.95KK92 pKa = 10.41DD93 pKa = 3.27GSFTEE98 pKa = 4.64LGDD101 pKa = 3.47VTRR104 pKa = 11.84GQGEE108 pKa = 4.23RR109 pKa = 11.84NDD111 pKa = 3.73LKK113 pKa = 10.84RR114 pKa = 11.84ACEE117 pKa = 4.11LIKK120 pKa = 10.36EE121 pKa = 4.5GKK123 pKa = 9.75SISEE127 pKa = 4.25VADD130 pKa = 3.65ACPTTYY136 pKa = 10.67VKK138 pKa = 10.68YY139 pKa = 10.69SRR141 pKa = 11.84GLRR144 pKa = 11.84EE145 pKa = 3.85LALVASEE152 pKa = 4.61AYY154 pKa = 8.84EE155 pKa = 4.17HH156 pKa = 6.56EE157 pKa = 4.61SVRR160 pKa = 11.84GVWIYY165 pKa = 10.95GQTGVGKK172 pKa = 9.75SHH174 pKa = 7.96LARR177 pKa = 11.84DD178 pKa = 4.24VYY180 pKa = 11.26PNLFDD185 pKa = 4.77KK186 pKa = 11.08SQSKK190 pKa = 7.92WWDD193 pKa = 4.15GYY195 pKa = 10.7CNEE198 pKa = 4.43HH199 pKa = 6.15TVLIDD204 pKa = 6.14DD205 pKa = 5.98LDD207 pKa = 4.16TDD209 pKa = 4.6CLGHH213 pKa = 5.94YY214 pKa = 9.58LKK216 pKa = 10.65RR217 pKa = 11.84WADD220 pKa = 3.39KK221 pKa = 10.05YY222 pKa = 11.24ACTGEE227 pKa = 4.49TKK229 pKa = 10.72GGTIKK234 pKa = 10.46LQHH237 pKa = 6.25RR238 pKa = 11.84VFVVTSNYY246 pKa = 10.07LPWEE250 pKa = 3.94LHH252 pKa = 6.33KK253 pKa = 11.15DD254 pKa = 3.5DD255 pKa = 5.79LMAEE259 pKa = 4.68AISRR263 pKa = 11.84RR264 pKa = 11.84FTIYY268 pKa = 10.36HH269 pKa = 6.51KK270 pKa = 7.8EE271 pKa = 4.01TRR273 pKa = 11.84DD274 pKa = 3.47QKK276 pKa = 11.63LEE278 pKa = 4.1DD279 pKa = 4.42CEE281 pKa = 4.32VLTYY285 pKa = 10.93ADD287 pKa = 3.6RR288 pKa = 11.84HH289 pKa = 5.75NIII292 pKa = 4.56
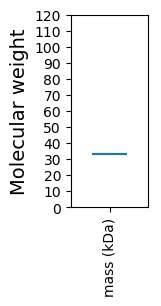
Molecular weight: 33.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141MJB3|A0A141MJB3_9VIRU ATP-dependent helicase Rep OS=Ctenophore-associated circular genome 1 OX=1778570 PE=3 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.5LSDD6 pKa = 3.51NRR8 pKa = 11.84VVNYY12 pKa = 9.86VFTWNNYY19 pKa = 7.47TDD21 pKa = 3.82DD22 pKa = 3.67VKK24 pKa = 11.58AFIGNFANDD33 pKa = 3.29YY34 pKa = 9.95CKK36 pKa = 9.84WLVYY40 pKa = 10.54GEE42 pKa = 4.34EE43 pKa = 4.37VGASGTPHH51 pKa = 6.46LQGCFSLYY59 pKa = 9.92DD60 pKa = 3.67KK61 pKa = 11.2KK62 pKa = 10.99RR63 pKa = 11.84ITQLCKK69 pKa = 10.58LGFKK73 pKa = 10.45AHH75 pKa = 7.0LEE77 pKa = 3.99PMKK80 pKa = 10.66GSPKK84 pKa = 9.96QAIAYY89 pKa = 7.12CKK91 pKa = 9.95KK92 pKa = 10.41DD93 pKa = 3.27GSFTEE98 pKa = 4.64LGDD101 pKa = 3.47VTRR104 pKa = 11.84GQGEE108 pKa = 4.23RR109 pKa = 11.84NDD111 pKa = 3.73LKK113 pKa = 10.84RR114 pKa = 11.84ACEE117 pKa = 4.11LIKK120 pKa = 10.36EE121 pKa = 4.5GKK123 pKa = 9.75SISEE127 pKa = 4.25VADD130 pKa = 3.65ACPTTYY136 pKa = 10.67VKK138 pKa = 10.68YY139 pKa = 10.69SRR141 pKa = 11.84GLRR144 pKa = 11.84EE145 pKa = 3.85LALVASEE152 pKa = 4.61AYY154 pKa = 8.84EE155 pKa = 4.17HH156 pKa = 6.56EE157 pKa = 4.61SVRR160 pKa = 11.84GVWIYY165 pKa = 10.95GQTGVGKK172 pKa = 9.75SHH174 pKa = 7.96LARR177 pKa = 11.84DD178 pKa = 4.24VYY180 pKa = 11.26PNLFDD185 pKa = 4.77KK186 pKa = 11.08SQSKK190 pKa = 7.92WWDD193 pKa = 4.15GYY195 pKa = 10.7CNEE198 pKa = 4.43HH199 pKa = 6.15TVLIDD204 pKa = 6.14DD205 pKa = 5.98LDD207 pKa = 4.16TDD209 pKa = 4.6CLGHH213 pKa = 5.94YY214 pKa = 9.58LKK216 pKa = 10.65RR217 pKa = 11.84WADD220 pKa = 3.39KK221 pKa = 10.05YY222 pKa = 11.24ACTGEE227 pKa = 4.49TKK229 pKa = 10.72GGTIKK234 pKa = 10.46LQHH237 pKa = 6.25RR238 pKa = 11.84VFVVTSNYY246 pKa = 10.07LPWEE250 pKa = 3.94LHH252 pKa = 6.33KK253 pKa = 11.15DD254 pKa = 3.5DD255 pKa = 5.79LMAEE259 pKa = 4.68AISRR263 pKa = 11.84RR264 pKa = 11.84FTIYY268 pKa = 10.36HH269 pKa = 6.51KK270 pKa = 7.8EE271 pKa = 4.01TRR273 pKa = 11.84DD274 pKa = 3.47QKK276 pKa = 11.63LEE278 pKa = 4.1DD279 pKa = 4.42CEE281 pKa = 4.32VLTYY285 pKa = 10.93ADD287 pKa = 3.6RR288 pKa = 11.84HH289 pKa = 5.75NIII292 pKa = 4.56
MM1 pKa = 7.84PKK3 pKa = 10.5LSDD6 pKa = 3.51NRR8 pKa = 11.84VVNYY12 pKa = 9.86VFTWNNYY19 pKa = 7.47TDD21 pKa = 3.82DD22 pKa = 3.67VKK24 pKa = 11.58AFIGNFANDD33 pKa = 3.29YY34 pKa = 9.95CKK36 pKa = 9.84WLVYY40 pKa = 10.54GEE42 pKa = 4.34EE43 pKa = 4.37VGASGTPHH51 pKa = 6.46LQGCFSLYY59 pKa = 9.92DD60 pKa = 3.67KK61 pKa = 11.2KK62 pKa = 10.99RR63 pKa = 11.84ITQLCKK69 pKa = 10.58LGFKK73 pKa = 10.45AHH75 pKa = 7.0LEE77 pKa = 3.99PMKK80 pKa = 10.66GSPKK84 pKa = 9.96QAIAYY89 pKa = 7.12CKK91 pKa = 9.95KK92 pKa = 10.41DD93 pKa = 3.27GSFTEE98 pKa = 4.64LGDD101 pKa = 3.47VTRR104 pKa = 11.84GQGEE108 pKa = 4.23RR109 pKa = 11.84NDD111 pKa = 3.73LKK113 pKa = 10.84RR114 pKa = 11.84ACEE117 pKa = 4.11LIKK120 pKa = 10.36EE121 pKa = 4.5GKK123 pKa = 9.75SISEE127 pKa = 4.25VADD130 pKa = 3.65ACPTTYY136 pKa = 10.67VKK138 pKa = 10.68YY139 pKa = 10.69SRR141 pKa = 11.84GLRR144 pKa = 11.84EE145 pKa = 3.85LALVASEE152 pKa = 4.61AYY154 pKa = 8.84EE155 pKa = 4.17HH156 pKa = 6.56EE157 pKa = 4.61SVRR160 pKa = 11.84GVWIYY165 pKa = 10.95GQTGVGKK172 pKa = 9.75SHH174 pKa = 7.96LARR177 pKa = 11.84DD178 pKa = 4.24VYY180 pKa = 11.26PNLFDD185 pKa = 4.77KK186 pKa = 11.08SQSKK190 pKa = 7.92WWDD193 pKa = 4.15GYY195 pKa = 10.7CNEE198 pKa = 4.43HH199 pKa = 6.15TVLIDD204 pKa = 6.14DD205 pKa = 5.98LDD207 pKa = 4.16TDD209 pKa = 4.6CLGHH213 pKa = 5.94YY214 pKa = 9.58LKK216 pKa = 10.65RR217 pKa = 11.84WADD220 pKa = 3.39KK221 pKa = 10.05YY222 pKa = 11.24ACTGEE227 pKa = 4.49TKK229 pKa = 10.72GGTIKK234 pKa = 10.46LQHH237 pKa = 6.25RR238 pKa = 11.84VFVVTSNYY246 pKa = 10.07LPWEE250 pKa = 3.94LHH252 pKa = 6.33KK253 pKa = 11.15DD254 pKa = 3.5DD255 pKa = 5.79LMAEE259 pKa = 4.68AISRR263 pKa = 11.84RR264 pKa = 11.84FTIYY268 pKa = 10.36HH269 pKa = 6.51KK270 pKa = 7.8EE271 pKa = 4.01TRR273 pKa = 11.84DD274 pKa = 3.47QKK276 pKa = 11.63LEE278 pKa = 4.1DD279 pKa = 4.42CEE281 pKa = 4.32VLTYY285 pKa = 10.93ADD287 pKa = 3.6RR288 pKa = 11.84HH289 pKa = 5.75NIII292 pKa = 4.56
Molecular weight: 33.44 kDa
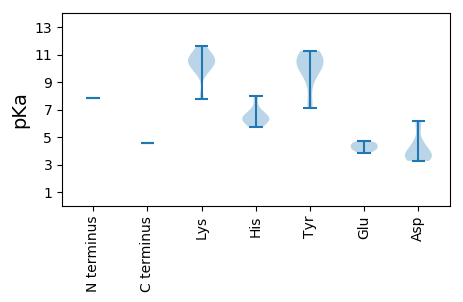
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
292 |
292 |
292 |
292.0 |
33.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.164 ± 0.0 | 3.425 ± 0.0 |
7.534 ± 0.0 | 6.507 ± 0.0 |
3.082 ± 0.0 | 7.534 ± 0.0 |
3.425 ± 0.0 | 4.11 ± 0.0 |
8.562 ± 0.0 | 8.562 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.027 ± 0.0 | 3.767 ± 0.0 |
2.397 ± 0.0 | 2.74 ± 0.0 |
5.137 ± 0.0 | 5.137 ± 0.0 |
6.164 ± 0.0 | 6.507 ± 0.0 |
2.397 ± 0.0 | 5.822 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
