
Wenzhou pacific spadenose shark paramyxovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Scoliodonvirus; Scoliodon scoliodonvirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
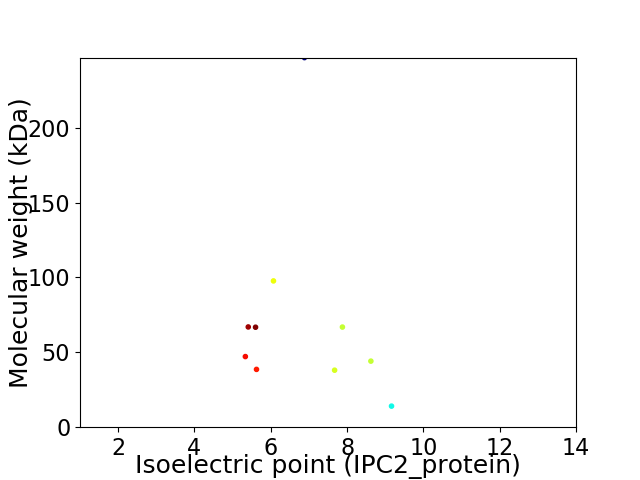
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
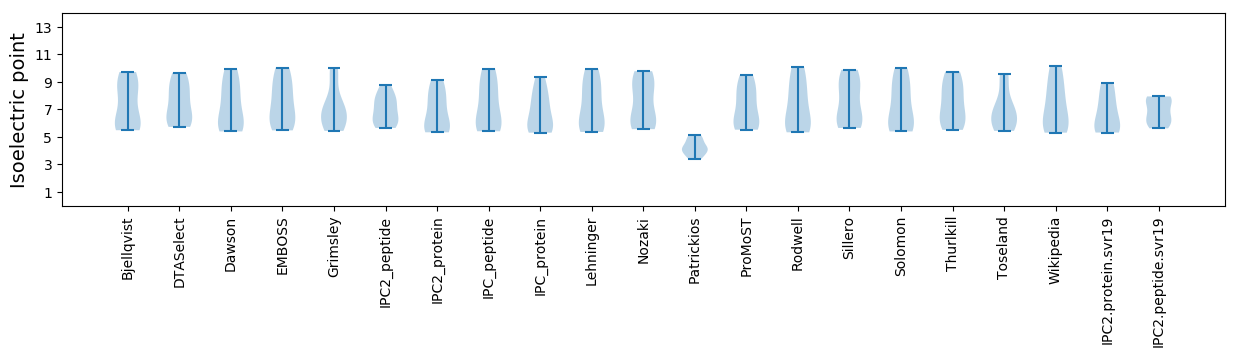
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GMX6|A0A2P1GMX6_9MONO Putative glycoprotein OS=Wenzhou pacific spadenose shark paramyxovirus OX=2116452 PE=4 SV=1
MM1 pKa = 7.75ALTFKK6 pKa = 10.56GIKK9 pKa = 9.41SGRR12 pKa = 11.84SGLGKK17 pKa = 10.11SGKK20 pKa = 10.32LSMDD24 pKa = 3.61EE25 pKa = 4.26LCEE28 pKa = 4.16LSCSPFVSSAANPAFGGEE46 pKa = 4.27GSRR49 pKa = 11.84VALISDD55 pKa = 4.1EE56 pKa = 4.46SVAAAPLFIYY66 pKa = 10.38PDD68 pKa = 3.97DD69 pKa = 4.31KK70 pKa = 11.57PEE72 pKa = 4.07EE73 pKa = 4.0FWVDD77 pKa = 3.0LFRR80 pKa = 11.84IFGATFGGGTLNEE93 pKa = 4.41VARR96 pKa = 11.84EE97 pKa = 3.92AALAAVLLSPFKK109 pKa = 11.1NGIDD113 pKa = 3.6LAKK116 pKa = 10.26EE117 pKa = 3.81VAKK120 pKa = 11.04NSGASPEE127 pKa = 4.1IVLLDD132 pKa = 3.86LSPTGRR138 pKa = 11.84IITRR142 pKa = 11.84DD143 pKa = 3.67PASKK147 pKa = 9.64HH148 pKa = 5.89APMLGDD154 pKa = 6.11LIDD157 pKa = 3.87QGDD160 pKa = 4.04ANLQSRR166 pKa = 11.84LFMAKK171 pKa = 9.34SWDD174 pKa = 3.7LDD176 pKa = 3.51QEE178 pKa = 4.31IPEE181 pKa = 4.1EE182 pKa = 4.58AEE184 pKa = 4.06LIRR187 pKa = 11.84AIFCQIYY194 pKa = 9.48GFSGKK199 pKa = 10.46AMANYY204 pKa = 9.68VEE206 pKa = 5.3AYY208 pKa = 10.47ASAKK212 pKa = 9.73LHH214 pKa = 5.98NKK216 pKa = 8.63VFYY219 pKa = 9.72TNRR222 pKa = 11.84FITNSLVITAHH233 pKa = 6.63GATQIVDD240 pKa = 4.14IIRR243 pKa = 11.84ASGQIRR249 pKa = 11.84HH250 pKa = 6.21LLVAEE255 pKa = 4.55ILASGASTGGGGAIGEE271 pKa = 4.81IIRR274 pKa = 11.84HH275 pKa = 5.16HH276 pKa = 6.35AVYY279 pKa = 10.32IAGTQMFQYY288 pKa = 10.37YY289 pKa = 7.59VTVQNVMKK297 pKa = 10.36SRR299 pKa = 11.84RR300 pKa = 11.84ALASISGAVGEE311 pKa = 4.42ARR313 pKa = 11.84TLIKK317 pKa = 10.61LMRR320 pKa = 11.84LQARR324 pKa = 11.84YY325 pKa = 8.87GALGPFVKK333 pKa = 10.4FLCLPDD339 pKa = 4.4HH340 pKa = 6.23NQFSGNAYY348 pKa = 9.23PSLFSMALGIEE359 pKa = 4.19DD360 pKa = 3.81VKK362 pKa = 11.43GKK364 pKa = 8.84TSHH367 pKa = 6.9KK368 pKa = 8.37YY369 pKa = 8.52VHH371 pKa = 7.3DD372 pKa = 3.45ISYY375 pKa = 10.78VNKK378 pKa = 10.26SLYY381 pKa = 10.33EE382 pKa = 3.85LGKK385 pKa = 10.36EE386 pKa = 3.56IGRR389 pKa = 11.84ASFLEE394 pKa = 4.01MDD396 pKa = 3.66QSALNEE402 pKa = 4.16LEE404 pKa = 4.29VTAEE408 pKa = 3.76EE409 pKa = 4.39AANMDD414 pKa = 3.96RR415 pKa = 11.84VNRR418 pKa = 11.84AALKK422 pKa = 10.85ALMDD426 pKa = 5.23DD427 pKa = 5.69DD428 pKa = 5.2YY429 pKa = 11.97DD430 pKa = 5.87SDD432 pKa = 3.91DD433 pKa = 4.18QEE435 pKa = 4.08II436 pKa = 3.74
MM1 pKa = 7.75ALTFKK6 pKa = 10.56GIKK9 pKa = 9.41SGRR12 pKa = 11.84SGLGKK17 pKa = 10.11SGKK20 pKa = 10.32LSMDD24 pKa = 3.61EE25 pKa = 4.26LCEE28 pKa = 4.16LSCSPFVSSAANPAFGGEE46 pKa = 4.27GSRR49 pKa = 11.84VALISDD55 pKa = 4.1EE56 pKa = 4.46SVAAAPLFIYY66 pKa = 10.38PDD68 pKa = 3.97DD69 pKa = 4.31KK70 pKa = 11.57PEE72 pKa = 4.07EE73 pKa = 4.0FWVDD77 pKa = 3.0LFRR80 pKa = 11.84IFGATFGGGTLNEE93 pKa = 4.41VARR96 pKa = 11.84EE97 pKa = 3.92AALAAVLLSPFKK109 pKa = 11.1NGIDD113 pKa = 3.6LAKK116 pKa = 10.26EE117 pKa = 3.81VAKK120 pKa = 11.04NSGASPEE127 pKa = 4.1IVLLDD132 pKa = 3.86LSPTGRR138 pKa = 11.84IITRR142 pKa = 11.84DD143 pKa = 3.67PASKK147 pKa = 9.64HH148 pKa = 5.89APMLGDD154 pKa = 6.11LIDD157 pKa = 3.87QGDD160 pKa = 4.04ANLQSRR166 pKa = 11.84LFMAKK171 pKa = 9.34SWDD174 pKa = 3.7LDD176 pKa = 3.51QEE178 pKa = 4.31IPEE181 pKa = 4.1EE182 pKa = 4.58AEE184 pKa = 4.06LIRR187 pKa = 11.84AIFCQIYY194 pKa = 9.48GFSGKK199 pKa = 10.46AMANYY204 pKa = 9.68VEE206 pKa = 5.3AYY208 pKa = 10.47ASAKK212 pKa = 9.73LHH214 pKa = 5.98NKK216 pKa = 8.63VFYY219 pKa = 9.72TNRR222 pKa = 11.84FITNSLVITAHH233 pKa = 6.63GATQIVDD240 pKa = 4.14IIRR243 pKa = 11.84ASGQIRR249 pKa = 11.84HH250 pKa = 6.21LLVAEE255 pKa = 4.55ILASGASTGGGGAIGEE271 pKa = 4.81IIRR274 pKa = 11.84HH275 pKa = 5.16HH276 pKa = 6.35AVYY279 pKa = 10.32IAGTQMFQYY288 pKa = 10.37YY289 pKa = 7.59VTVQNVMKK297 pKa = 10.36SRR299 pKa = 11.84RR300 pKa = 11.84ALASISGAVGEE311 pKa = 4.42ARR313 pKa = 11.84TLIKK317 pKa = 10.61LMRR320 pKa = 11.84LQARR324 pKa = 11.84YY325 pKa = 8.87GALGPFVKK333 pKa = 10.4FLCLPDD339 pKa = 4.4HH340 pKa = 6.23NQFSGNAYY348 pKa = 9.23PSLFSMALGIEE359 pKa = 4.19DD360 pKa = 3.81VKK362 pKa = 11.43GKK364 pKa = 8.84TSHH367 pKa = 6.9KK368 pKa = 8.37YY369 pKa = 8.52VHH371 pKa = 7.3DD372 pKa = 3.45ISYY375 pKa = 10.78VNKK378 pKa = 10.26SLYY381 pKa = 10.33EE382 pKa = 3.85LGKK385 pKa = 10.36EE386 pKa = 3.56IGRR389 pKa = 11.84ASFLEE394 pKa = 4.01MDD396 pKa = 3.66QSALNEE402 pKa = 4.16LEE404 pKa = 4.29VTAEE408 pKa = 3.76EE409 pKa = 4.39AANMDD414 pKa = 3.96RR415 pKa = 11.84VNRR418 pKa = 11.84AALKK422 pKa = 10.85ALMDD426 pKa = 5.23DD427 pKa = 5.69DD428 pKa = 5.2YY429 pKa = 11.97DD430 pKa = 5.87SDD432 pKa = 3.91DD433 pKa = 4.18QEE435 pKa = 4.08II436 pKa = 3.74
Molecular weight: 47.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GMY3|A0A2P1GMY3_9MONO Hemagglutinin-neuraminidase OS=Wenzhou pacific spadenose shark paramyxovirus OX=2116452 PE=4 SV=1
MM1 pKa = 7.24TSVSGSKK8 pKa = 9.97PLWRR12 pKa = 11.84RR13 pKa = 11.84FLSSALRR20 pKa = 11.84SLRR23 pKa = 11.84TRR25 pKa = 11.84SFRR28 pKa = 11.84YY29 pKa = 8.01RR30 pKa = 11.84TLSQTSQSSDD40 pKa = 3.14PRR42 pKa = 11.84HH43 pKa = 6.46LPTPPHH49 pKa = 6.89PSPTSSPEE57 pKa = 3.61TSTEE61 pKa = 4.07SIYY64 pKa = 11.23CVMNPLSAPLPCEE77 pKa = 3.38TWTEE81 pKa = 3.43QDD83 pKa = 3.15TYY85 pKa = 8.65KK86 pKa = 10.67TSWIQPQCQGRR97 pKa = 11.84GRR99 pKa = 11.84KK100 pKa = 8.6FSKK103 pKa = 9.21GTPVTYY109 pKa = 10.86IMTDD113 pKa = 2.74KK114 pKa = 10.95GLEE117 pKa = 3.86EE118 pKa = 4.18ARR120 pKa = 11.84TYY122 pKa = 10.27TLL124 pKa = 3.85
MM1 pKa = 7.24TSVSGSKK8 pKa = 9.97PLWRR12 pKa = 11.84RR13 pKa = 11.84FLSSALRR20 pKa = 11.84SLRR23 pKa = 11.84TRR25 pKa = 11.84SFRR28 pKa = 11.84YY29 pKa = 8.01RR30 pKa = 11.84TLSQTSQSSDD40 pKa = 3.14PRR42 pKa = 11.84HH43 pKa = 6.46LPTPPHH49 pKa = 6.89PSPTSSPEE57 pKa = 3.61TSTEE61 pKa = 4.07SIYY64 pKa = 11.23CVMNPLSAPLPCEE77 pKa = 3.38TWTEE81 pKa = 3.43QDD83 pKa = 3.15TYY85 pKa = 8.65KK86 pKa = 10.67TSWIQPQCQGRR97 pKa = 11.84GRR99 pKa = 11.84KK100 pKa = 8.6FSKK103 pKa = 9.21GTPVTYY109 pKa = 10.86IMTDD113 pKa = 2.74KK114 pKa = 10.95GLEE117 pKa = 3.86EE118 pKa = 4.18ARR120 pKa = 11.84TYY122 pKa = 10.27TLL124 pKa = 3.85
Molecular weight: 14.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6480 |
124 |
2164 |
648.0 |
72.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.648 ± 0.556 | 1.59 ± 0.23 |
5.247 ± 0.448 | 5.88 ± 0.67 |
3.565 ± 0.377 | 5.741 ± 0.295 |
2.361 ± 0.163 | 7.33 ± 0.587 |
5.556 ± 0.989 | 10.309 ± 0.602 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.167 | 3.704 ± 0.153 |
5.664 ± 0.631 | 4.028 ± 0.273 |
5.37 ± 0.446 | 8.812 ± 0.503 |
6.404 ± 0.394 | 5.802 ± 0.431 |
1.019 ± 0.169 | 3.58 ± 0.222 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
