
Rahnella aquatilis (strain ATCC 33071 / DSM 4594 / JCM 1683 / NBRC 105701 / NCIMB 13365 / CIP 78.65)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Yersiniaceae; Rahnella; Rahnella aquatilis
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
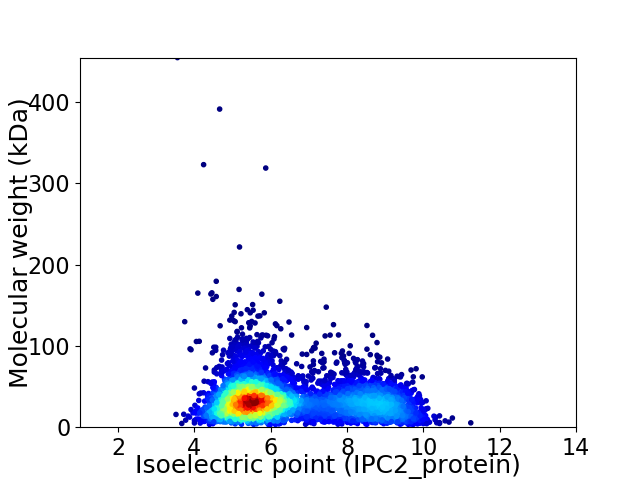
Virtual 2D-PAGE plot for 4841 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H2IQ67|H2IQ67_RAHAC Alkylated DNA repair protein OS=Rahnella aquatilis (strain ATCC 33071 / DSM 4594 / JCM 1683 / NBRC 105701 / NCIMB 13365 / CIP 78.65) OX=745277 GN=Rahaq2_3719 PE=4 SV=1
MM1 pKa = 7.33VFNKK5 pKa = 8.67ITTCLFLAFNLLPLADD21 pKa = 3.53VVYY24 pKa = 11.09AEE26 pKa = 4.68TLPACSGFQITEE38 pKa = 4.18SCSATPTTLEE48 pKa = 3.87NKK50 pKa = 8.54TLNTYY55 pKa = 8.29TVADD59 pKa = 3.99GAEE62 pKa = 4.2MQFSGLTVSGDD73 pKa = 3.27SGGAFHH79 pKa = 7.44LGNGSTLEE87 pKa = 4.31IIPEE91 pKa = 4.05NTSGYY96 pKa = 10.5SEE98 pKa = 4.14FTGNSSSGNGGAIFAKK114 pKa = 10.25EE115 pKa = 3.67NSTVYY120 pKa = 10.86LEE122 pKa = 4.09NTLFIDD128 pKa = 5.04NVAHH132 pKa = 7.51DD133 pKa = 4.6YY134 pKa = 11.44GGAIYY139 pKa = 8.71MYY141 pKa = 10.33GSNDD145 pKa = 3.5EE146 pKa = 4.39GDD148 pKa = 3.57VDD150 pKa = 4.13LTITNAVFSGNIASEE165 pKa = 4.41GKK167 pKa = 10.48GGAIYY172 pKa = 10.64SLNNKK177 pKa = 8.14VQLTNILFEE186 pKa = 4.53NNQAKK191 pKa = 9.74TNGNGSNGNGSGGAIDD207 pKa = 4.28ATDD210 pKa = 3.52NTTDD214 pKa = 2.84SHH216 pKa = 7.92GSMFITNSEE225 pKa = 4.07FTGNQAEE232 pKa = 4.53GQGGAIYY239 pKa = 9.7TNSASTPFFVDD250 pKa = 2.98INIDD254 pKa = 3.5EE255 pKa = 4.53NYY257 pKa = 10.21EE258 pKa = 4.15DD259 pKa = 4.14NNGVAYY265 pKa = 10.82YY266 pKa = 10.15HH267 pKa = 7.53DD268 pKa = 4.31NVADD272 pKa = 3.95NYY274 pKa = 11.55GNDD277 pKa = 3.52PDD279 pKa = 4.04AAGGGFMYY287 pKa = 10.53LGHH290 pKa = 6.27SEE292 pKa = 4.44ANFDD296 pKa = 3.35IAADD300 pKa = 3.57KK301 pKa = 10.64TLVIGNTANDD311 pKa = 3.86GAYY314 pKa = 10.52DD315 pKa = 4.16SIAGYY320 pKa = 9.84GAIFKK325 pKa = 10.38RR326 pKa = 11.84GAGEE330 pKa = 4.05LVLNADD336 pKa = 3.38NSGFTGVFDD345 pKa = 3.67ILEE348 pKa = 4.38GTVTLGRR355 pKa = 11.84DD356 pKa = 3.43DD357 pKa = 3.91TLVNVGDD364 pKa = 4.21TNCQSEE370 pKa = 4.5ADD372 pKa = 3.77TCHH375 pKa = 7.14GIVLGNANSMNDD387 pKa = 3.46KK388 pKa = 10.83AILNVGSTQQTFKK401 pKa = 10.85NAFMGHH407 pKa = 5.85EE408 pKa = 4.01NSTLNIDD415 pKa = 3.21AGGNVIVNSGYY426 pKa = 9.25MAGTTTGAGDD436 pKa = 3.51LTVAEE441 pKa = 4.69NGSFTLTGAQSMGLDD456 pKa = 3.16GDD458 pKa = 3.91IVVEE462 pKa = 4.11QNAVLSVQGDD472 pKa = 3.96AEE474 pKa = 4.31DD475 pKa = 4.4LAILQADD482 pKa = 3.94PQSIVLDD489 pKa = 3.92GGVLDD494 pKa = 5.08LSDD497 pKa = 3.29MSTFNGQTNQVNDD510 pKa = 3.64GLTITGTGGTVIGNDD525 pKa = 3.56DD526 pKa = 4.71LVTLGAGTDD535 pKa = 3.46YY536 pKa = 11.05HH537 pKa = 8.55IGDD540 pKa = 4.09GTSAADD546 pKa = 3.5GVYY549 pKa = 10.35VVIDD553 pKa = 3.85AGDD556 pKa = 3.67GRR558 pKa = 11.84VTLADD563 pKa = 3.34NNAYY567 pKa = 10.69LGTTQIASGTLQITDD582 pKa = 3.38NSQLGDD588 pKa = 3.29KK589 pKa = 10.37AYY591 pKa = 10.43NRR593 pKa = 11.84QVIFTDD599 pKa = 3.98PAQASTLEE607 pKa = 4.1VSAPADD613 pKa = 3.2GSGQVDD619 pKa = 3.61TTSAQTGKK627 pKa = 10.57ARR629 pKa = 11.84DD630 pKa = 3.32IEE632 pKa = 4.33MRR634 pKa = 11.84ASGTMQVDD642 pKa = 4.25DD643 pKa = 4.86GVTTQWGGLMADD655 pKa = 3.65STGAQADD662 pKa = 3.97VNSTFTKK669 pKa = 9.66TGNGTLILDD678 pKa = 4.21GTGSSHH684 pKa = 5.82SAVRR688 pKa = 11.84VEE690 pKa = 4.37QGTLQGGAEE699 pKa = 4.31NIIASASSLYY709 pKa = 11.07VGDD712 pKa = 3.57NATFEE717 pKa = 4.4TGLDD721 pKa = 3.51QQVQSLDD728 pKa = 3.54VASGGTIVISDD739 pKa = 4.04DD740 pKa = 3.63TQLMLMEE747 pKa = 4.76QDD749 pKa = 2.95TSAALDD755 pKa = 3.42ASLFADD761 pKa = 3.47TDD763 pKa = 3.84GTLVNATSGMSLQGTLNTNMTLDD786 pKa = 3.77ALTYY790 pKa = 10.59LPGVTVNGSLDD801 pKa = 3.55NSAGTASLQNGTAGDD816 pKa = 3.98TLTVNGDD823 pKa = 3.41YY824 pKa = 10.78TGGGTVVLEE833 pKa = 4.3TEE835 pKa = 4.41MNGDD839 pKa = 3.5TSATDD844 pKa = 3.46QLILNGNTSGDD855 pKa = 3.35TALAIVPVSGIGQATQAGIKK875 pKa = 10.22VVDD878 pKa = 5.3FVADD882 pKa = 3.63PQGNNNAADD891 pKa = 3.59FHH893 pKa = 6.84LAGDD897 pKa = 3.59QQYY900 pKa = 10.68INMGAYY906 pKa = 9.94DD907 pKa = 4.0YY908 pKa = 11.82SLVQDD913 pKa = 3.6NQDD916 pKa = 2.57WYY918 pKa = 11.36LRR920 pKa = 11.84SQVVTPDD927 pKa = 3.72PQPEE931 pKa = 4.14PVPGGDD937 pKa = 4.56DD938 pKa = 3.56FTPVLNPKK946 pKa = 9.07MGSYY950 pKa = 9.73MNNLRR955 pKa = 11.84IANNVFSMTAQDD967 pKa = 3.87HH968 pKa = 6.67AGADD972 pKa = 3.65HH973 pKa = 7.8DD974 pKa = 4.29NLKK977 pKa = 10.7LRR979 pKa = 11.84VDD981 pKa = 4.04TARR984 pKa = 11.84DD985 pKa = 3.16SATFGNQIDD994 pKa = 4.03SHH996 pKa = 7.76NDD998 pKa = 2.85TSVAQLSANFLQHH1011 pKa = 6.37PAGNGEE1017 pKa = 4.2LLAGIVAGYY1026 pKa = 9.96SNNSGDD1032 pKa = 3.87STSVLTGRR1040 pKa = 11.84KK1041 pKa = 9.08SDD1043 pKa = 4.0NDD1045 pKa = 3.68SQGYY1049 pKa = 9.57AVGLTGAWYY1058 pKa = 10.32QDD1060 pKa = 3.36AQAHH1064 pKa = 4.91QGAYY1068 pKa = 10.02LDD1070 pKa = 3.91SWLQYY1075 pKa = 10.66AWFDD1079 pKa = 3.54NSVSEE1084 pKa = 5.06EE1085 pKa = 4.01DD1086 pKa = 3.55TGDD1089 pKa = 3.92DD1090 pKa = 3.68NYY1092 pKa = 11.08HH1093 pKa = 6.67ASGLLASLEE1102 pKa = 3.86GGYY1105 pKa = 10.26RR1106 pKa = 11.84FTLMQGNQWNWTLQPQAQVIYY1127 pKa = 10.54QGVKK1131 pKa = 10.06QKK1133 pKa = 11.25DD1134 pKa = 3.7FTSASQSKK1142 pKa = 7.7VTQTGDD1148 pKa = 3.72DD1149 pKa = 4.08NIQTRR1154 pKa = 11.84LGLRR1158 pKa = 11.84SEE1160 pKa = 4.14WDD1162 pKa = 3.0IHH1164 pKa = 5.98PVSTLHH1170 pKa = 6.41IKK1172 pKa = 10.56PFVEE1176 pKa = 3.34ANYY1179 pKa = 10.2RR1180 pKa = 11.84HH1181 pKa = 6.11NSEE1184 pKa = 4.09QTSLNVDD1191 pKa = 3.41GVNFTDD1197 pKa = 3.7NTAKK1201 pKa = 10.76DD1202 pKa = 3.62SAEE1205 pKa = 4.03LSTGISGNLGANTTVWGKK1223 pKa = 10.44VGQQEE1228 pKa = 4.36GSDD1231 pKa = 3.9DD1232 pKa = 3.89FSQTEE1237 pKa = 3.97ATVGVSVRR1245 pKa = 11.84WW1246 pKa = 3.4
MM1 pKa = 7.33VFNKK5 pKa = 8.67ITTCLFLAFNLLPLADD21 pKa = 3.53VVYY24 pKa = 11.09AEE26 pKa = 4.68TLPACSGFQITEE38 pKa = 4.18SCSATPTTLEE48 pKa = 3.87NKK50 pKa = 8.54TLNTYY55 pKa = 8.29TVADD59 pKa = 3.99GAEE62 pKa = 4.2MQFSGLTVSGDD73 pKa = 3.27SGGAFHH79 pKa = 7.44LGNGSTLEE87 pKa = 4.31IIPEE91 pKa = 4.05NTSGYY96 pKa = 10.5SEE98 pKa = 4.14FTGNSSSGNGGAIFAKK114 pKa = 10.25EE115 pKa = 3.67NSTVYY120 pKa = 10.86LEE122 pKa = 4.09NTLFIDD128 pKa = 5.04NVAHH132 pKa = 7.51DD133 pKa = 4.6YY134 pKa = 11.44GGAIYY139 pKa = 8.71MYY141 pKa = 10.33GSNDD145 pKa = 3.5EE146 pKa = 4.39GDD148 pKa = 3.57VDD150 pKa = 4.13LTITNAVFSGNIASEE165 pKa = 4.41GKK167 pKa = 10.48GGAIYY172 pKa = 10.64SLNNKK177 pKa = 8.14VQLTNILFEE186 pKa = 4.53NNQAKK191 pKa = 9.74TNGNGSNGNGSGGAIDD207 pKa = 4.28ATDD210 pKa = 3.52NTTDD214 pKa = 2.84SHH216 pKa = 7.92GSMFITNSEE225 pKa = 4.07FTGNQAEE232 pKa = 4.53GQGGAIYY239 pKa = 9.7TNSASTPFFVDD250 pKa = 2.98INIDD254 pKa = 3.5EE255 pKa = 4.53NYY257 pKa = 10.21EE258 pKa = 4.15DD259 pKa = 4.14NNGVAYY265 pKa = 10.82YY266 pKa = 10.15HH267 pKa = 7.53DD268 pKa = 4.31NVADD272 pKa = 3.95NYY274 pKa = 11.55GNDD277 pKa = 3.52PDD279 pKa = 4.04AAGGGFMYY287 pKa = 10.53LGHH290 pKa = 6.27SEE292 pKa = 4.44ANFDD296 pKa = 3.35IAADD300 pKa = 3.57KK301 pKa = 10.64TLVIGNTANDD311 pKa = 3.86GAYY314 pKa = 10.52DD315 pKa = 4.16SIAGYY320 pKa = 9.84GAIFKK325 pKa = 10.38RR326 pKa = 11.84GAGEE330 pKa = 4.05LVLNADD336 pKa = 3.38NSGFTGVFDD345 pKa = 3.67ILEE348 pKa = 4.38GTVTLGRR355 pKa = 11.84DD356 pKa = 3.43DD357 pKa = 3.91TLVNVGDD364 pKa = 4.21TNCQSEE370 pKa = 4.5ADD372 pKa = 3.77TCHH375 pKa = 7.14GIVLGNANSMNDD387 pKa = 3.46KK388 pKa = 10.83AILNVGSTQQTFKK401 pKa = 10.85NAFMGHH407 pKa = 5.85EE408 pKa = 4.01NSTLNIDD415 pKa = 3.21AGGNVIVNSGYY426 pKa = 9.25MAGTTTGAGDD436 pKa = 3.51LTVAEE441 pKa = 4.69NGSFTLTGAQSMGLDD456 pKa = 3.16GDD458 pKa = 3.91IVVEE462 pKa = 4.11QNAVLSVQGDD472 pKa = 3.96AEE474 pKa = 4.31DD475 pKa = 4.4LAILQADD482 pKa = 3.94PQSIVLDD489 pKa = 3.92GGVLDD494 pKa = 5.08LSDD497 pKa = 3.29MSTFNGQTNQVNDD510 pKa = 3.64GLTITGTGGTVIGNDD525 pKa = 3.56DD526 pKa = 4.71LVTLGAGTDD535 pKa = 3.46YY536 pKa = 11.05HH537 pKa = 8.55IGDD540 pKa = 4.09GTSAADD546 pKa = 3.5GVYY549 pKa = 10.35VVIDD553 pKa = 3.85AGDD556 pKa = 3.67GRR558 pKa = 11.84VTLADD563 pKa = 3.34NNAYY567 pKa = 10.69LGTTQIASGTLQITDD582 pKa = 3.38NSQLGDD588 pKa = 3.29KK589 pKa = 10.37AYY591 pKa = 10.43NRR593 pKa = 11.84QVIFTDD599 pKa = 3.98PAQASTLEE607 pKa = 4.1VSAPADD613 pKa = 3.2GSGQVDD619 pKa = 3.61TTSAQTGKK627 pKa = 10.57ARR629 pKa = 11.84DD630 pKa = 3.32IEE632 pKa = 4.33MRR634 pKa = 11.84ASGTMQVDD642 pKa = 4.25DD643 pKa = 4.86GVTTQWGGLMADD655 pKa = 3.65STGAQADD662 pKa = 3.97VNSTFTKK669 pKa = 9.66TGNGTLILDD678 pKa = 4.21GTGSSHH684 pKa = 5.82SAVRR688 pKa = 11.84VEE690 pKa = 4.37QGTLQGGAEE699 pKa = 4.31NIIASASSLYY709 pKa = 11.07VGDD712 pKa = 3.57NATFEE717 pKa = 4.4TGLDD721 pKa = 3.51QQVQSLDD728 pKa = 3.54VASGGTIVISDD739 pKa = 4.04DD740 pKa = 3.63TQLMLMEE747 pKa = 4.76QDD749 pKa = 2.95TSAALDD755 pKa = 3.42ASLFADD761 pKa = 3.47TDD763 pKa = 3.84GTLVNATSGMSLQGTLNTNMTLDD786 pKa = 3.77ALTYY790 pKa = 10.59LPGVTVNGSLDD801 pKa = 3.55NSAGTASLQNGTAGDD816 pKa = 3.98TLTVNGDD823 pKa = 3.41YY824 pKa = 10.78TGGGTVVLEE833 pKa = 4.3TEE835 pKa = 4.41MNGDD839 pKa = 3.5TSATDD844 pKa = 3.46QLILNGNTSGDD855 pKa = 3.35TALAIVPVSGIGQATQAGIKK875 pKa = 10.22VVDD878 pKa = 5.3FVADD882 pKa = 3.63PQGNNNAADD891 pKa = 3.59FHH893 pKa = 6.84LAGDD897 pKa = 3.59QQYY900 pKa = 10.68INMGAYY906 pKa = 9.94DD907 pKa = 4.0YY908 pKa = 11.82SLVQDD913 pKa = 3.6NQDD916 pKa = 2.57WYY918 pKa = 11.36LRR920 pKa = 11.84SQVVTPDD927 pKa = 3.72PQPEE931 pKa = 4.14PVPGGDD937 pKa = 4.56DD938 pKa = 3.56FTPVLNPKK946 pKa = 9.07MGSYY950 pKa = 9.73MNNLRR955 pKa = 11.84IANNVFSMTAQDD967 pKa = 3.87HH968 pKa = 6.67AGADD972 pKa = 3.65HH973 pKa = 7.8DD974 pKa = 4.29NLKK977 pKa = 10.7LRR979 pKa = 11.84VDD981 pKa = 4.04TARR984 pKa = 11.84DD985 pKa = 3.16SATFGNQIDD994 pKa = 4.03SHH996 pKa = 7.76NDD998 pKa = 2.85TSVAQLSANFLQHH1011 pKa = 6.37PAGNGEE1017 pKa = 4.2LLAGIVAGYY1026 pKa = 9.96SNNSGDD1032 pKa = 3.87STSVLTGRR1040 pKa = 11.84KK1041 pKa = 9.08SDD1043 pKa = 4.0NDD1045 pKa = 3.68SQGYY1049 pKa = 9.57AVGLTGAWYY1058 pKa = 10.32QDD1060 pKa = 3.36AQAHH1064 pKa = 4.91QGAYY1068 pKa = 10.02LDD1070 pKa = 3.91SWLQYY1075 pKa = 10.66AWFDD1079 pKa = 3.54NSVSEE1084 pKa = 5.06EE1085 pKa = 4.01DD1086 pKa = 3.55TGDD1089 pKa = 3.92DD1090 pKa = 3.68NYY1092 pKa = 11.08HH1093 pKa = 6.67ASGLLASLEE1102 pKa = 3.86GGYY1105 pKa = 10.26RR1106 pKa = 11.84FTLMQGNQWNWTLQPQAQVIYY1127 pKa = 10.54QGVKK1131 pKa = 10.06QKK1133 pKa = 11.25DD1134 pKa = 3.7FTSASQSKK1142 pKa = 7.7VTQTGDD1148 pKa = 3.72DD1149 pKa = 4.08NIQTRR1154 pKa = 11.84LGLRR1158 pKa = 11.84SEE1160 pKa = 4.14WDD1162 pKa = 3.0IHH1164 pKa = 5.98PVSTLHH1170 pKa = 6.41IKK1172 pKa = 10.56PFVEE1176 pKa = 3.34ANYY1179 pKa = 10.2RR1180 pKa = 11.84HH1181 pKa = 6.11NSEE1184 pKa = 4.09QTSLNVDD1191 pKa = 3.41GVNFTDD1197 pKa = 3.7NTAKK1201 pKa = 10.76DD1202 pKa = 3.62SAEE1205 pKa = 4.03LSTGISGNLGANTTVWGKK1223 pKa = 10.44VGQQEE1228 pKa = 4.36GSDD1231 pKa = 3.9DD1232 pKa = 3.89FSQTEE1237 pKa = 3.97ATVGVSVRR1245 pKa = 11.84WW1246 pKa = 3.4
Molecular weight: 129.84 kDa
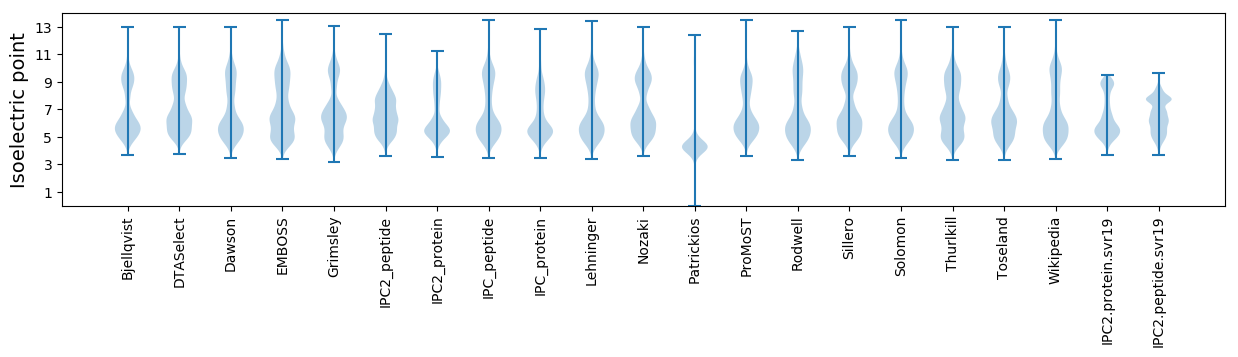
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H2IZ43|H2IZ43_RAHAC Uncharacterized protein OS=Rahnella aquatilis (strain ATCC 33071 / DSM 4594 / JCM 1683 / NBRC 105701 / NCIMB 13365 / CIP 78.65) OX=745277 GN=Rahaq2_1064 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.55GRR39 pKa = 11.84TRR41 pKa = 11.84LSASKK46 pKa = 10.74
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.55GRR39 pKa = 11.84TRR41 pKa = 11.84LSASKK46 pKa = 10.74
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1570583 |
23 |
4595 |
324.4 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.661 ± 0.042 | 1.032 ± 0.013 |
5.26 ± 0.024 | 5.44 ± 0.03 |
3.927 ± 0.026 | 7.491 ± 0.035 |
2.239 ± 0.02 | 5.808 ± 0.034 |
4.291 ± 0.026 | 10.812 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.752 ± 0.02 | 3.785 ± 0.031 |
4.559 ± 0.023 | 4.51 ± 0.034 |
5.39 ± 0.035 | 6.257 ± 0.03 |
5.483 ± 0.042 | 7.035 ± 0.029 |
1.417 ± 0.016 | 2.851 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
