
Human papillomavirus 179
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 15
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
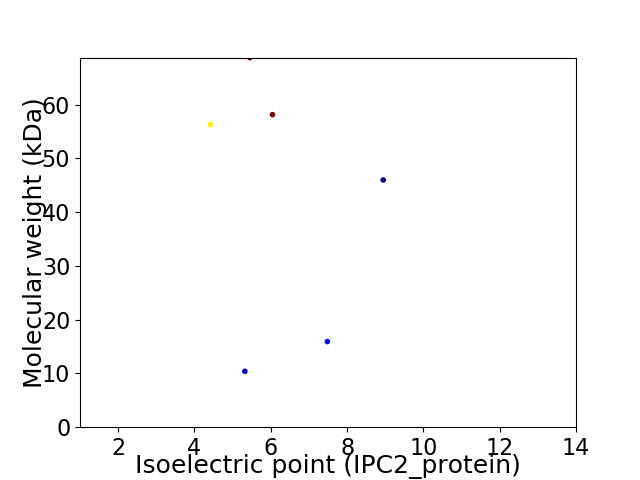
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S6FWD1|S6FWD1_9PAPI Protein E6 OS=Human papillomavirus 179 OX=1472342 GN=E6 PE=3 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84QKK8 pKa = 10.19RR9 pKa = 11.84ASINDD14 pKa = 3.96LYY16 pKa = 11.18RR17 pKa = 11.84NCRR20 pKa = 11.84LGGDD24 pKa = 3.81CPEE27 pKa = 4.16DD28 pKa = 3.14VKK30 pKa = 11.65NKK32 pKa = 10.14VEE34 pKa = 4.15GTTLADD40 pKa = 3.54RR41 pKa = 11.84LLQIFGSILYY51 pKa = 9.61FGNLGIGTGKK61 pKa = 8.83GTGGFGGYY69 pKa = 9.82KK70 pKa = 10.04PIGGTASKK78 pKa = 10.6GPEE81 pKa = 3.7ITVSRR86 pKa = 11.84PNVPIDD92 pKa = 3.62PLGGADD98 pKa = 5.25VIPLDD103 pKa = 4.57IINPEE108 pKa = 4.09APSVIPLQEE117 pKa = 4.55GGLPNIPVTDD127 pKa = 3.65TGITTTDD134 pKa = 2.94IAEE137 pKa = 4.05IDD139 pKa = 4.09IITTTNPIDD148 pKa = 3.83NPITTDD154 pKa = 3.39TQPTVITQNNDD165 pKa = 2.19IFIVDD170 pKa = 4.27MQPGPPPPKK179 pKa = 10.09RR180 pKa = 11.84IALDD184 pKa = 3.23VGTRR188 pKa = 11.84PFADD192 pKa = 2.81IEE194 pKa = 4.64LNVFRR199 pKa = 11.84EE200 pKa = 3.94PHH202 pKa = 6.22FDD204 pKa = 3.54SNVNVFVDD212 pKa = 3.84PNITGDD218 pKa = 3.72VVGLEE223 pKa = 4.19EE224 pKa = 5.69IEE226 pKa = 4.51LGPLNEE232 pKa = 4.1VAEE235 pKa = 4.29FAIEE239 pKa = 3.98EE240 pKa = 4.36GAGPSTSTPTQSIEE254 pKa = 4.07RR255 pKa = 11.84IINQGQRR262 pKa = 11.84LYY264 pKa = 11.36NRR266 pKa = 11.84FVQQVQTRR274 pKa = 11.84NPDD277 pKa = 3.43FVTKK281 pKa = 9.96PSRR284 pKa = 11.84LVTFEE289 pKa = 4.25FEE291 pKa = 4.6NPAFDD296 pKa = 5.93DD297 pKa = 4.59DD298 pKa = 3.99VTLTFEE304 pKa = 4.47QDD306 pKa = 3.23INDD309 pKa = 3.4VAAAPDD315 pKa = 3.7YY316 pKa = 10.31EE317 pKa = 4.12FRR319 pKa = 11.84DD320 pKa = 3.59IVRR323 pKa = 11.84LEE325 pKa = 3.92RR326 pKa = 11.84PIFTTTDD333 pKa = 2.64AGLRR337 pKa = 11.84LSRR340 pKa = 11.84LGQRR344 pKa = 11.84GSITTRR350 pKa = 11.84SGLTIGPRR358 pKa = 11.84VHH360 pKa = 7.14FYY362 pKa = 11.42YY363 pKa = 10.64DD364 pKa = 3.42FSEE367 pKa = 4.18ILPEE371 pKa = 3.89EE372 pKa = 4.5SIEE375 pKa = 4.08LSTFAIHH382 pKa = 6.89SNEE385 pKa = 4.26DD386 pKa = 3.37VFIDD390 pKa = 3.85PMAEE394 pKa = 3.9SSIVNSDD401 pKa = 2.76IGRR404 pKa = 11.84NPTFSDD410 pKa = 4.78DD411 pKa = 3.64YY412 pKa = 11.55LEE414 pKa = 6.45DD415 pKa = 3.41IFTEE419 pKa = 4.22NFNNAHH425 pKa = 6.3LVLTFGEE432 pKa = 4.01RR433 pKa = 11.84DD434 pKa = 3.47IIEE437 pKa = 4.28VPISVPKK444 pKa = 10.64AGINVFITGNSNTTVVDD461 pKa = 3.45YY462 pKa = 10.55TSIKK466 pKa = 8.27EE467 pKa = 4.25TTIITPSTNLINLKK481 pKa = 9.7PALGLYY487 pKa = 9.23TLGEE491 pKa = 4.38DD492 pKa = 5.57FIFDD496 pKa = 3.7PDD498 pKa = 3.78LFRR501 pKa = 11.84KK502 pKa = 9.07RR503 pKa = 11.84RR504 pKa = 11.84KK505 pKa = 9.5RR506 pKa = 11.84KK507 pKa = 9.64YY508 pKa = 10.39SDD510 pKa = 3.05VV511 pKa = 3.5
MM1 pKa = 7.45LRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84QKK8 pKa = 10.19RR9 pKa = 11.84ASINDD14 pKa = 3.96LYY16 pKa = 11.18RR17 pKa = 11.84NCRR20 pKa = 11.84LGGDD24 pKa = 3.81CPEE27 pKa = 4.16DD28 pKa = 3.14VKK30 pKa = 11.65NKK32 pKa = 10.14VEE34 pKa = 4.15GTTLADD40 pKa = 3.54RR41 pKa = 11.84LLQIFGSILYY51 pKa = 9.61FGNLGIGTGKK61 pKa = 8.83GTGGFGGYY69 pKa = 9.82KK70 pKa = 10.04PIGGTASKK78 pKa = 10.6GPEE81 pKa = 3.7ITVSRR86 pKa = 11.84PNVPIDD92 pKa = 3.62PLGGADD98 pKa = 5.25VIPLDD103 pKa = 4.57IINPEE108 pKa = 4.09APSVIPLQEE117 pKa = 4.55GGLPNIPVTDD127 pKa = 3.65TGITTTDD134 pKa = 2.94IAEE137 pKa = 4.05IDD139 pKa = 4.09IITTTNPIDD148 pKa = 3.83NPITTDD154 pKa = 3.39TQPTVITQNNDD165 pKa = 2.19IFIVDD170 pKa = 4.27MQPGPPPPKK179 pKa = 10.09RR180 pKa = 11.84IALDD184 pKa = 3.23VGTRR188 pKa = 11.84PFADD192 pKa = 2.81IEE194 pKa = 4.64LNVFRR199 pKa = 11.84EE200 pKa = 3.94PHH202 pKa = 6.22FDD204 pKa = 3.54SNVNVFVDD212 pKa = 3.84PNITGDD218 pKa = 3.72VVGLEE223 pKa = 4.19EE224 pKa = 5.69IEE226 pKa = 4.51LGPLNEE232 pKa = 4.1VAEE235 pKa = 4.29FAIEE239 pKa = 3.98EE240 pKa = 4.36GAGPSTSTPTQSIEE254 pKa = 4.07RR255 pKa = 11.84IINQGQRR262 pKa = 11.84LYY264 pKa = 11.36NRR266 pKa = 11.84FVQQVQTRR274 pKa = 11.84NPDD277 pKa = 3.43FVTKK281 pKa = 9.96PSRR284 pKa = 11.84LVTFEE289 pKa = 4.25FEE291 pKa = 4.6NPAFDD296 pKa = 5.93DD297 pKa = 4.59DD298 pKa = 3.99VTLTFEE304 pKa = 4.47QDD306 pKa = 3.23INDD309 pKa = 3.4VAAAPDD315 pKa = 3.7YY316 pKa = 10.31EE317 pKa = 4.12FRR319 pKa = 11.84DD320 pKa = 3.59IVRR323 pKa = 11.84LEE325 pKa = 3.92RR326 pKa = 11.84PIFTTTDD333 pKa = 2.64AGLRR337 pKa = 11.84LSRR340 pKa = 11.84LGQRR344 pKa = 11.84GSITTRR350 pKa = 11.84SGLTIGPRR358 pKa = 11.84VHH360 pKa = 7.14FYY362 pKa = 11.42YY363 pKa = 10.64DD364 pKa = 3.42FSEE367 pKa = 4.18ILPEE371 pKa = 3.89EE372 pKa = 4.5SIEE375 pKa = 4.08LSTFAIHH382 pKa = 6.89SNEE385 pKa = 4.26DD386 pKa = 3.37VFIDD390 pKa = 3.85PMAEE394 pKa = 3.9SSIVNSDD401 pKa = 2.76IGRR404 pKa = 11.84NPTFSDD410 pKa = 4.78DD411 pKa = 3.64YY412 pKa = 11.55LEE414 pKa = 6.45DD415 pKa = 3.41IFTEE419 pKa = 4.22NFNNAHH425 pKa = 6.3LVLTFGEE432 pKa = 4.01RR433 pKa = 11.84DD434 pKa = 3.47IIEE437 pKa = 4.28VPISVPKK444 pKa = 10.64AGINVFITGNSNTTVVDD461 pKa = 3.45YY462 pKa = 10.55TSIKK466 pKa = 8.27EE467 pKa = 4.25TTIITPSTNLINLKK481 pKa = 9.7PALGLYY487 pKa = 9.23TLGEE491 pKa = 4.38DD492 pKa = 5.57FIFDD496 pKa = 3.7PDD498 pKa = 3.78LFRR501 pKa = 11.84KK502 pKa = 9.07RR503 pKa = 11.84RR504 pKa = 11.84KK505 pKa = 9.5RR506 pKa = 11.84KK507 pKa = 9.64YY508 pKa = 10.39SDD510 pKa = 3.05VV511 pKa = 3.5
Molecular weight: 56.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S6G386|S6G386_9PAPI Regulatory protein E2 OS=Human papillomavirus 179 OX=1472342 GN=E2 PE=3 SV=1
MM1 pKa = 6.99NQADD5 pKa = 3.75LTEE8 pKa = 5.0RR9 pKa = 11.84FDD11 pKa = 4.39ALQDD15 pKa = 3.62QLLTLYY21 pKa = 7.6EE22 pKa = 4.17TAPTDD27 pKa = 3.25IQSQITHH34 pKa = 6.52WALTRR39 pKa = 11.84RR40 pKa = 11.84LNVLMYY46 pKa = 8.72YY47 pKa = 10.37ARR49 pKa = 11.84QEE51 pKa = 4.31GYY53 pKa = 10.83RR54 pKa = 11.84NLGLQALPTLVVSEE68 pKa = 4.63YY69 pKa = 9.55NAKK72 pKa = 10.17VAIKK76 pKa = 10.34MMMLLKK82 pKa = 10.46SLSEE86 pKa = 4.03SQYY89 pKa = 11.39GKK91 pKa = 10.6EE92 pKa = 3.82PWTLADD98 pKa = 4.24ASAEE102 pKa = 4.03LVLTSPKK109 pKa = 9.21HH110 pKa = 4.43TFKK113 pKa = 10.89KK114 pKa = 10.26GAFQVEE120 pKa = 4.66VYY122 pKa = 10.82YY123 pKa = 11.15DD124 pKa = 4.0NDD126 pKa = 3.67PQNANVYY133 pKa = 7.16TQWEE137 pKa = 4.37HH138 pKa = 7.41IYY140 pKa = 11.22YY141 pKa = 10.43EE142 pKa = 4.3DD143 pKa = 6.11LNDD146 pKa = 3.23IWHH149 pKa = 6.65KK150 pKa = 10.9VPGDD154 pKa = 3.42VDD156 pKa = 4.19HH157 pKa = 7.16NGLSFTDD164 pKa = 3.43VTGDD168 pKa = 3.08KK169 pKa = 11.02NYY171 pKa = 10.58FLLFHH176 pKa = 7.39EE177 pKa = 4.89DD178 pKa = 3.0AQRR181 pKa = 11.84YY182 pKa = 9.24SNTGQWTVKK191 pKa = 10.2FKK193 pKa = 10.42STTISSVVTSSRR205 pKa = 11.84QQSSNQKK212 pKa = 9.45RR213 pKa = 11.84DD214 pKa = 3.37QQSRR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84DD221 pKa = 3.38SSPEE225 pKa = 3.82EE226 pKa = 3.89GTSRR230 pKa = 11.84AFRR233 pKa = 11.84RR234 pKa = 11.84SEE236 pKa = 3.84RR237 pKa = 11.84DD238 pKa = 3.24PEE240 pKa = 4.08EE241 pKa = 4.89SISTTTTSPTTTRR254 pKa = 11.84QRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84GEE263 pKa = 3.99RR264 pKa = 11.84GDD266 pKa = 3.51QQGEE270 pKa = 4.42STPEE274 pKa = 3.39RR275 pKa = 11.84PRR277 pKa = 11.84AKK279 pKa = 9.93RR280 pKa = 11.84SRR282 pKa = 11.84GSSPVSPEE290 pKa = 3.87EE291 pKa = 3.52VGSRR295 pKa = 11.84HH296 pKa = 5.92RR297 pKa = 11.84SVPKK301 pKa = 9.76HH302 pKa = 4.78HH303 pKa = 7.18LGRR306 pKa = 11.84LRR308 pKa = 11.84RR309 pKa = 11.84LQEE312 pKa = 3.79EE313 pKa = 4.14ARR315 pKa = 11.84DD316 pKa = 3.93PPVLCIKK323 pKa = 10.78GPANNLKK330 pKa = 10.12CWRR333 pKa = 11.84NRR335 pKa = 11.84FNVRR339 pKa = 11.84FASLFFKK346 pKa = 10.79ASSVFKK352 pKa = 10.47WLGDD356 pKa = 3.59TDD358 pKa = 4.86CPTNTSRR365 pKa = 11.84MLIAFHH371 pKa = 6.52SLNEE375 pKa = 3.96RR376 pKa = 11.84SRR378 pKa = 11.84FLQLVTLPKK387 pKa = 9.4GTSYY391 pKa = 11.27SYY393 pKa = 11.76GSLDD397 pKa = 3.5SLL399 pKa = 4.67
MM1 pKa = 6.99NQADD5 pKa = 3.75LTEE8 pKa = 5.0RR9 pKa = 11.84FDD11 pKa = 4.39ALQDD15 pKa = 3.62QLLTLYY21 pKa = 7.6EE22 pKa = 4.17TAPTDD27 pKa = 3.25IQSQITHH34 pKa = 6.52WALTRR39 pKa = 11.84RR40 pKa = 11.84LNVLMYY46 pKa = 8.72YY47 pKa = 10.37ARR49 pKa = 11.84QEE51 pKa = 4.31GYY53 pKa = 10.83RR54 pKa = 11.84NLGLQALPTLVVSEE68 pKa = 4.63YY69 pKa = 9.55NAKK72 pKa = 10.17VAIKK76 pKa = 10.34MMMLLKK82 pKa = 10.46SLSEE86 pKa = 4.03SQYY89 pKa = 11.39GKK91 pKa = 10.6EE92 pKa = 3.82PWTLADD98 pKa = 4.24ASAEE102 pKa = 4.03LVLTSPKK109 pKa = 9.21HH110 pKa = 4.43TFKK113 pKa = 10.89KK114 pKa = 10.26GAFQVEE120 pKa = 4.66VYY122 pKa = 10.82YY123 pKa = 11.15DD124 pKa = 4.0NDD126 pKa = 3.67PQNANVYY133 pKa = 7.16TQWEE137 pKa = 4.37HH138 pKa = 7.41IYY140 pKa = 11.22YY141 pKa = 10.43EE142 pKa = 4.3DD143 pKa = 6.11LNDD146 pKa = 3.23IWHH149 pKa = 6.65KK150 pKa = 10.9VPGDD154 pKa = 3.42VDD156 pKa = 4.19HH157 pKa = 7.16NGLSFTDD164 pKa = 3.43VTGDD168 pKa = 3.08KK169 pKa = 11.02NYY171 pKa = 10.58FLLFHH176 pKa = 7.39EE177 pKa = 4.89DD178 pKa = 3.0AQRR181 pKa = 11.84YY182 pKa = 9.24SNTGQWTVKK191 pKa = 10.2FKK193 pKa = 10.42STTISSVVTSSRR205 pKa = 11.84QQSSNQKK212 pKa = 9.45RR213 pKa = 11.84DD214 pKa = 3.37QQSRR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84DD221 pKa = 3.38SSPEE225 pKa = 3.82EE226 pKa = 3.89GTSRR230 pKa = 11.84AFRR233 pKa = 11.84RR234 pKa = 11.84SEE236 pKa = 3.84RR237 pKa = 11.84DD238 pKa = 3.24PEE240 pKa = 4.08EE241 pKa = 4.89SISTTTTSPTTTRR254 pKa = 11.84QRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84GEE263 pKa = 3.99RR264 pKa = 11.84GDD266 pKa = 3.51QQGEE270 pKa = 4.42STPEE274 pKa = 3.39RR275 pKa = 11.84PRR277 pKa = 11.84AKK279 pKa = 9.93RR280 pKa = 11.84SRR282 pKa = 11.84GSSPVSPEE290 pKa = 3.87EE291 pKa = 3.52VGSRR295 pKa = 11.84HH296 pKa = 5.92RR297 pKa = 11.84SVPKK301 pKa = 9.76HH302 pKa = 4.78HH303 pKa = 7.18LGRR306 pKa = 11.84LRR308 pKa = 11.84RR309 pKa = 11.84LQEE312 pKa = 3.79EE313 pKa = 4.14ARR315 pKa = 11.84DD316 pKa = 3.93PPVLCIKK323 pKa = 10.78GPANNLKK330 pKa = 10.12CWRR333 pKa = 11.84NRR335 pKa = 11.84FNVRR339 pKa = 11.84FASLFFKK346 pKa = 10.79ASSVFKK352 pKa = 10.47WLGDD356 pKa = 3.59TDD358 pKa = 4.86CPTNTSRR365 pKa = 11.84MLIAFHH371 pKa = 6.52SLNEE375 pKa = 3.96RR376 pKa = 11.84SRR378 pKa = 11.84FLQLVTLPKK387 pKa = 9.4GTSYY391 pKa = 11.27SYY393 pKa = 11.76GSLDD397 pKa = 3.5SLL399 pKa = 4.67
Molecular weight: 46.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2257 |
95 |
602 |
376.2 |
42.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.937 ± 0.733 | 2.171 ± 0.907 |
6.823 ± 0.525 | 5.76 ± 0.354 |
4.962 ± 0.417 | 5.405 ± 0.817 |
1.994 ± 0.332 | 6.07 ± 1.316 |
4.829 ± 0.631 | 8.994 ± 0.626 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.639 ± 0.351 | 5.76 ± 0.338 |
5.804 ± 1.086 | 3.81 ± 0.548 |
6.336 ± 0.895 | 6.868 ± 0.849 |
6.602 ± 0.943 | 5.716 ± 0.306 |
1.063 ± 0.302 | 3.456 ± 0.52 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
