
Knoellia subterranea KCTC 19937
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Intrasporangiaceae; Knoellia; Knoellia subterranea
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
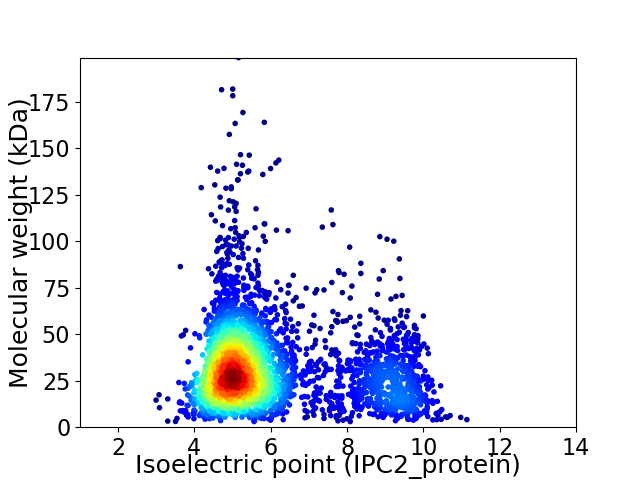
Virtual 2D-PAGE plot for 3418 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0JJN4|A0A0A0JJN4_9MICO Uncharacterized protein OS=Knoellia subterranea KCTC 19937 OX=1385521 GN=N803_14245 PE=4 SV=1
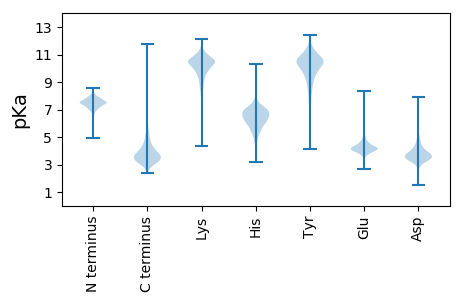
MM1 pKa = 7.22IAVAALVVGLIAVAISSPAADD22 pKa = 3.33VRR24 pKa = 11.84GHH26 pKa = 4.82VCRR29 pKa = 11.84AVADD33 pKa = 4.01VFNLDD38 pKa = 3.93ASCSGGGDD46 pKa = 3.52DD47 pKa = 6.36GEE49 pKa = 4.87TEE51 pKa = 4.2TGDD54 pKa = 3.39PDD56 pKa = 4.51QEE58 pKa = 4.4FEE60 pKa = 4.14PEE62 pKa = 3.6KK63 pKa = 10.96CKK65 pKa = 10.28IHH67 pKa = 5.54EE68 pKa = 4.58TGDD71 pKa = 3.59EE72 pKa = 4.11YY73 pKa = 11.78SSVIKK78 pKa = 10.49IGFIEE83 pKa = 4.09IGEE86 pKa = 4.15NAGFVVTEE94 pKa = 4.14YY95 pKa = 11.48SDD97 pKa = 3.64GTVTMMATNGSEE109 pKa = 3.61IGATGGFGADD119 pKa = 2.92AAYY122 pKa = 10.55GEE124 pKa = 4.75LEE126 pKa = 4.3ANASVDD132 pKa = 3.86FGGGLKK138 pKa = 9.85FDD140 pKa = 4.89YY141 pKa = 10.93GSTWTFEE148 pKa = 4.31SQEE151 pKa = 3.95QADD154 pKa = 4.3EE155 pKa = 4.27FRR157 pKa = 11.84EE158 pKa = 3.98QLDD161 pKa = 4.69DD162 pKa = 3.84YY163 pKa = 12.03LMDD166 pKa = 3.58QWSMTHH172 pKa = 6.28PVCGPTGMCIPRR184 pKa = 11.84PTTGAEE190 pKa = 4.03PPPVPSTTFGGIALTGEE207 pKa = 4.18VSADD211 pKa = 3.39LGISLTSGTTPLTQAEE227 pKa = 4.37LTQQGVAGSISGDD240 pKa = 3.47AHH242 pKa = 4.53WTVTNDD248 pKa = 3.86NNNTPDD254 pKa = 4.52DD255 pKa = 4.65PSDD258 pKa = 3.49DD259 pKa = 3.25TRR261 pKa = 11.84TYY263 pKa = 8.75VTDD266 pKa = 3.28MSLNSEE272 pKa = 4.14VTGQIGLATGGVGSMVGMSYY292 pKa = 10.45SIKK295 pKa = 10.31KK296 pKa = 10.11DD297 pKa = 2.92ADD299 pKa = 3.51GNIIEE304 pKa = 4.24VQIVSTSQVDD314 pKa = 3.56SSGGAGAEE322 pKa = 4.19GEE324 pKa = 4.51VNQGQGKK331 pKa = 7.83NQNSGGGSVTDD342 pKa = 3.56TTTDD346 pKa = 2.91GDD348 pKa = 4.29VVVTEE353 pKa = 4.23TTLAIDD359 pKa = 4.64PDD361 pKa = 4.39DD362 pKa = 4.28SASQQIVQDD371 pKa = 3.49WMGGDD376 pKa = 3.75GDD378 pKa = 3.86FAYY381 pKa = 9.94PGLIKK386 pKa = 10.83LNMVDD391 pKa = 5.03PSQADD396 pKa = 3.61PNDD399 pKa = 3.73PFAMLLHH406 pKa = 6.43NNATSSTIAYY416 pKa = 9.37DD417 pKa = 3.28HH418 pKa = 6.64VSDD421 pKa = 4.84VEE423 pKa = 4.56GFALNVKK430 pKa = 10.18FGLAFGVDD438 pKa = 4.05FSMEE442 pKa = 3.98NSEE445 pKa = 4.3TTATEE450 pKa = 3.78ATYY453 pKa = 10.84LGAPRR458 pKa = 11.84PDD460 pKa = 3.11GTRR463 pKa = 11.84PVLDD467 pKa = 3.42YY468 pKa = 10.09TEE470 pKa = 4.78CVSS473 pKa = 3.84
MM1 pKa = 7.22IAVAALVVGLIAVAISSPAADD22 pKa = 3.33VRR24 pKa = 11.84GHH26 pKa = 4.82VCRR29 pKa = 11.84AVADD33 pKa = 4.01VFNLDD38 pKa = 3.93ASCSGGGDD46 pKa = 3.52DD47 pKa = 6.36GEE49 pKa = 4.87TEE51 pKa = 4.2TGDD54 pKa = 3.39PDD56 pKa = 4.51QEE58 pKa = 4.4FEE60 pKa = 4.14PEE62 pKa = 3.6KK63 pKa = 10.96CKK65 pKa = 10.28IHH67 pKa = 5.54EE68 pKa = 4.58TGDD71 pKa = 3.59EE72 pKa = 4.11YY73 pKa = 11.78SSVIKK78 pKa = 10.49IGFIEE83 pKa = 4.09IGEE86 pKa = 4.15NAGFVVTEE94 pKa = 4.14YY95 pKa = 11.48SDD97 pKa = 3.64GTVTMMATNGSEE109 pKa = 3.61IGATGGFGADD119 pKa = 2.92AAYY122 pKa = 10.55GEE124 pKa = 4.75LEE126 pKa = 4.3ANASVDD132 pKa = 3.86FGGGLKK138 pKa = 9.85FDD140 pKa = 4.89YY141 pKa = 10.93GSTWTFEE148 pKa = 4.31SQEE151 pKa = 3.95QADD154 pKa = 4.3EE155 pKa = 4.27FRR157 pKa = 11.84EE158 pKa = 3.98QLDD161 pKa = 4.69DD162 pKa = 3.84YY163 pKa = 12.03LMDD166 pKa = 3.58QWSMTHH172 pKa = 6.28PVCGPTGMCIPRR184 pKa = 11.84PTTGAEE190 pKa = 4.03PPPVPSTTFGGIALTGEE207 pKa = 4.18VSADD211 pKa = 3.39LGISLTSGTTPLTQAEE227 pKa = 4.37LTQQGVAGSISGDD240 pKa = 3.47AHH242 pKa = 4.53WTVTNDD248 pKa = 3.86NNNTPDD254 pKa = 4.52DD255 pKa = 4.65PSDD258 pKa = 3.49DD259 pKa = 3.25TRR261 pKa = 11.84TYY263 pKa = 8.75VTDD266 pKa = 3.28MSLNSEE272 pKa = 4.14VTGQIGLATGGVGSMVGMSYY292 pKa = 10.45SIKK295 pKa = 10.31KK296 pKa = 10.11DD297 pKa = 2.92ADD299 pKa = 3.51GNIIEE304 pKa = 4.24VQIVSTSQVDD314 pKa = 3.56SSGGAGAEE322 pKa = 4.19GEE324 pKa = 4.51VNQGQGKK331 pKa = 7.83NQNSGGGSVTDD342 pKa = 3.56TTTDD346 pKa = 2.91GDD348 pKa = 4.29VVVTEE353 pKa = 4.23TTLAIDD359 pKa = 4.64PDD361 pKa = 4.39DD362 pKa = 4.28SASQQIVQDD371 pKa = 3.49WMGGDD376 pKa = 3.75GDD378 pKa = 3.86FAYY381 pKa = 9.94PGLIKK386 pKa = 10.83LNMVDD391 pKa = 5.03PSQADD396 pKa = 3.61PNDD399 pKa = 3.73PFAMLLHH406 pKa = 6.43NNATSSTIAYY416 pKa = 9.37DD417 pKa = 3.28HH418 pKa = 6.64VSDD421 pKa = 4.84VEE423 pKa = 4.56GFALNVKK430 pKa = 10.18FGLAFGVDD438 pKa = 4.05FSMEE442 pKa = 3.98NSEE445 pKa = 4.3TTATEE450 pKa = 3.78ATYY453 pKa = 10.84LGAPRR458 pKa = 11.84PDD460 pKa = 3.11GTRR463 pKa = 11.84PVLDD467 pKa = 3.42YY468 pKa = 10.09TEE470 pKa = 4.78CVSS473 pKa = 3.84
Molecular weight: 49.04 kDa
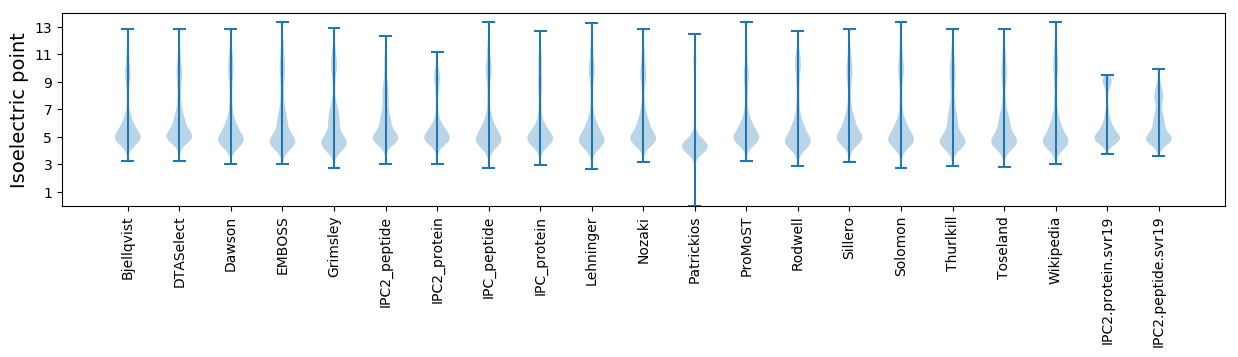
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0JM32|A0A0A0JM32_9MICO GGACT domain-containing protein OS=Knoellia subterranea KCTC 19937 OX=1385521 GN=N803_14590 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1069712 |
29 |
1816 |
313.0 |
33.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.99 ± 0.053 | 0.639 ± 0.009 |
6.408 ± 0.037 | 5.762 ± 0.039 |
2.827 ± 0.026 | 9.245 ± 0.038 |
2.212 ± 0.02 | 3.896 ± 0.033 |
2.213 ± 0.035 | 9.949 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.909 ± 0.019 | 1.895 ± 0.023 |
5.447 ± 0.032 | 2.675 ± 0.023 |
7.348 ± 0.042 | 5.619 ± 0.029 |
6.378 ± 0.035 | 9.229 ± 0.043 |
1.562 ± 0.016 | 1.797 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
