
Mink circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
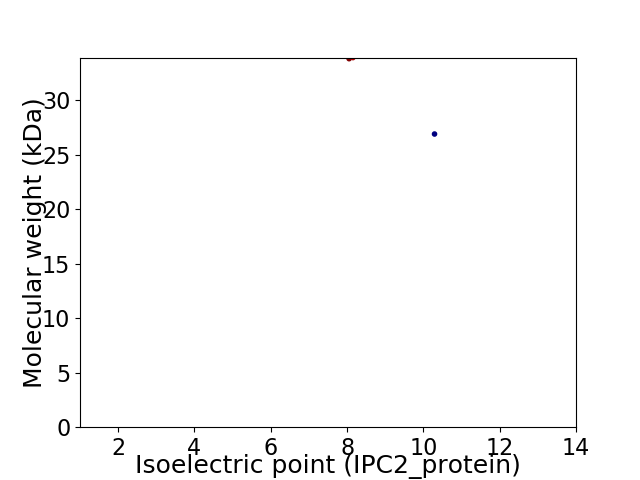
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
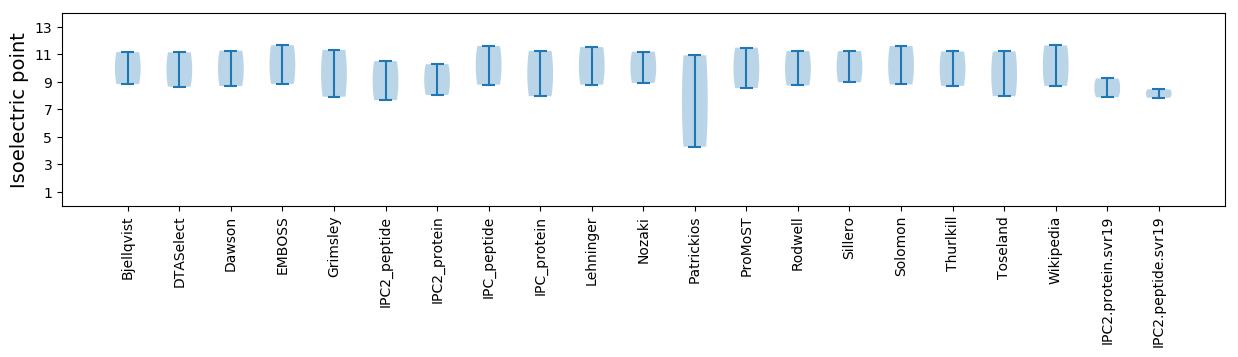
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8SQV8|W8SQV8_9CIRC Cap protein OS=Mink circovirus OX=1475143 GN=Cap PE=3 SV=1
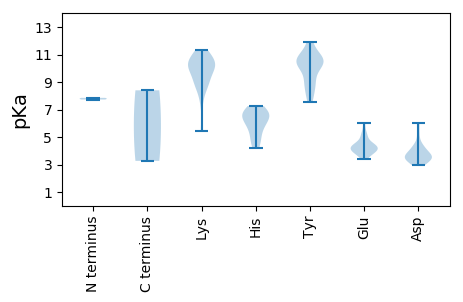
MM1 pKa = 7.85IMAGHH6 pKa = 6.6PCKK9 pKa = 10.35RR10 pKa = 11.84YY11 pKa = 10.21CFTINNYY18 pKa = 10.01LPEE21 pKa = 6.0DD22 pKa = 3.57EE23 pKa = 5.18AAVKK27 pKa = 10.46EE28 pKa = 4.22FLTEE32 pKa = 4.13ANCVYY37 pKa = 10.74AVVGKK42 pKa = 9.99EE43 pKa = 3.89VGEE46 pKa = 4.31SGTPHH51 pKa = 6.55LQGFCNLKK59 pKa = 10.0KK60 pKa = 10.55KK61 pKa = 9.69MRR63 pKa = 11.84FEE65 pKa = 3.98PFKK68 pKa = 10.66RR69 pKa = 11.84AIGGRR74 pKa = 11.84AHH76 pKa = 7.25IEE78 pKa = 3.77QSRR81 pKa = 11.84GTDD84 pKa = 2.97VDD86 pKa = 3.53NKK88 pKa = 10.23RR89 pKa = 11.84YY90 pKa = 9.37CSKK93 pKa = 11.18GGDD96 pKa = 3.5LLLEE100 pKa = 4.34VGEE103 pKa = 4.99PSAQGKK109 pKa = 10.21RR110 pKa = 11.84SDD112 pKa = 3.64LKK114 pKa = 11.13EE115 pKa = 3.98AVTLLNNGGTMTDD128 pKa = 3.17VARR131 pKa = 11.84AHH133 pKa = 7.0PEE135 pKa = 3.52TFIRR139 pKa = 11.84YY140 pKa = 8.49GRR142 pKa = 11.84GLRR145 pKa = 11.84DD146 pKa = 3.4YY147 pKa = 10.63VIQAGLTKK155 pKa = 9.92PRR157 pKa = 11.84AWKK160 pKa = 8.99TEE162 pKa = 3.42VHH164 pKa = 6.47VIVGPPGVGKK174 pKa = 10.24SRR176 pKa = 11.84HH177 pKa = 4.21VQEE180 pKa = 4.47TAGEE184 pKa = 3.95NALYY188 pKa = 8.75WKK190 pKa = 10.03PRR192 pKa = 11.84GKK194 pKa = 9.01WWDD197 pKa = 3.55GYY199 pKa = 8.86TGQSHH204 pKa = 6.09VVLDD208 pKa = 5.2DD209 pKa = 4.05FYY211 pKa = 11.9GWLPYY216 pKa = 10.72DD217 pKa = 4.39DD218 pKa = 5.99LLRR221 pKa = 11.84LCDD224 pKa = 4.13RR225 pKa = 11.84YY226 pKa = 10.18PLRR229 pKa = 11.84VEE231 pKa = 4.35TKK233 pKa = 8.08GGTVEE238 pKa = 5.39FVAKK242 pKa = 9.8VIWITSNKK250 pKa = 7.52QVKK253 pKa = 9.23DD254 pKa = 3.0WYY256 pKa = 10.23DD257 pKa = 3.67YY258 pKa = 11.91EE259 pKa = 4.14EE260 pKa = 4.86LKK262 pKa = 10.89VDD264 pKa = 3.46ARR266 pKa = 11.84ALYY269 pKa = 10.23RR270 pKa = 11.84RR271 pKa = 11.84LTTYY275 pKa = 10.56QVMRR279 pKa = 11.84QGGEE283 pKa = 3.99LYY285 pKa = 10.04NVQMTGDD292 pKa = 3.53NKK294 pKa = 10.49INYY297 pKa = 8.4
MM1 pKa = 7.85IMAGHH6 pKa = 6.6PCKK9 pKa = 10.35RR10 pKa = 11.84YY11 pKa = 10.21CFTINNYY18 pKa = 10.01LPEE21 pKa = 6.0DD22 pKa = 3.57EE23 pKa = 5.18AAVKK27 pKa = 10.46EE28 pKa = 4.22FLTEE32 pKa = 4.13ANCVYY37 pKa = 10.74AVVGKK42 pKa = 9.99EE43 pKa = 3.89VGEE46 pKa = 4.31SGTPHH51 pKa = 6.55LQGFCNLKK59 pKa = 10.0KK60 pKa = 10.55KK61 pKa = 9.69MRR63 pKa = 11.84FEE65 pKa = 3.98PFKK68 pKa = 10.66RR69 pKa = 11.84AIGGRR74 pKa = 11.84AHH76 pKa = 7.25IEE78 pKa = 3.77QSRR81 pKa = 11.84GTDD84 pKa = 2.97VDD86 pKa = 3.53NKK88 pKa = 10.23RR89 pKa = 11.84YY90 pKa = 9.37CSKK93 pKa = 11.18GGDD96 pKa = 3.5LLLEE100 pKa = 4.34VGEE103 pKa = 4.99PSAQGKK109 pKa = 10.21RR110 pKa = 11.84SDD112 pKa = 3.64LKK114 pKa = 11.13EE115 pKa = 3.98AVTLLNNGGTMTDD128 pKa = 3.17VARR131 pKa = 11.84AHH133 pKa = 7.0PEE135 pKa = 3.52TFIRR139 pKa = 11.84YY140 pKa = 8.49GRR142 pKa = 11.84GLRR145 pKa = 11.84DD146 pKa = 3.4YY147 pKa = 10.63VIQAGLTKK155 pKa = 9.92PRR157 pKa = 11.84AWKK160 pKa = 8.99TEE162 pKa = 3.42VHH164 pKa = 6.47VIVGPPGVGKK174 pKa = 10.24SRR176 pKa = 11.84HH177 pKa = 4.21VQEE180 pKa = 4.47TAGEE184 pKa = 3.95NALYY188 pKa = 8.75WKK190 pKa = 10.03PRR192 pKa = 11.84GKK194 pKa = 9.01WWDD197 pKa = 3.55GYY199 pKa = 8.86TGQSHH204 pKa = 6.09VVLDD208 pKa = 5.2DD209 pKa = 4.05FYY211 pKa = 11.9GWLPYY216 pKa = 10.72DD217 pKa = 4.39DD218 pKa = 5.99LLRR221 pKa = 11.84LCDD224 pKa = 4.13RR225 pKa = 11.84YY226 pKa = 10.18PLRR229 pKa = 11.84VEE231 pKa = 4.35TKK233 pKa = 8.08GGTVEE238 pKa = 5.39FVAKK242 pKa = 9.8VIWITSNKK250 pKa = 7.52QVKK253 pKa = 9.23DD254 pKa = 3.0WYY256 pKa = 10.23DD257 pKa = 3.67YY258 pKa = 11.91EE259 pKa = 4.14EE260 pKa = 4.86LKK262 pKa = 10.89VDD264 pKa = 3.46ARR266 pKa = 11.84ALYY269 pKa = 10.23RR270 pKa = 11.84RR271 pKa = 11.84LTTYY275 pKa = 10.56QVMRR279 pKa = 11.84QGGEE283 pKa = 3.99LYY285 pKa = 10.04NVQMTGDD292 pKa = 3.53NKK294 pKa = 10.49INYY297 pKa = 8.4
Molecular weight: 33.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8SQV8|W8SQV8_9CIRC Cap protein OS=Mink circovirus OX=1475143 GN=Cap PE=3 SV=1
MM1 pKa = 7.73PVRR4 pKa = 11.84SRR6 pKa = 11.84YY7 pKa = 7.8SRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WRR14 pKa = 11.84RR15 pKa = 11.84NRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84GPQRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.46ARR28 pKa = 11.84GGYY31 pKa = 8.14RR32 pKa = 11.84WRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 10.0NGIFNARR43 pKa = 11.84LQTEE47 pKa = 4.15EE48 pKa = 4.3TLTVASAATPSWCVGHH64 pKa = 6.73VNVTLKK70 pKa = 11.3SMMPKK75 pKa = 10.66GPGTTTNLYY84 pKa = 10.72GMPFSYY90 pKa = 10.84YY91 pKa = 10.36RR92 pKa = 11.84IRR94 pKa = 11.84KK95 pKa = 8.94LKK97 pKa = 10.45IEE99 pKa = 4.52FLPQIGITSNRR110 pKa = 11.84SYY112 pKa = 11.36GGTAIILDD120 pKa = 3.65GDD122 pKa = 4.31YY123 pKa = 10.79ISEE126 pKa = 4.45GKK128 pKa = 8.66NQTYY132 pKa = 10.76DD133 pKa = 3.31PLANHH138 pKa = 6.51SSRR141 pKa = 11.84HH142 pKa = 4.93NLTNIHH148 pKa = 5.45RR149 pKa = 11.84HH150 pKa = 4.68SRR152 pKa = 11.84YY153 pKa = 7.57FTPKK157 pKa = 8.81PQHH160 pKa = 5.74SSKK163 pKa = 11.04NISTYY168 pKa = 9.15FQPNNKK174 pKa = 8.63QLQEE178 pKa = 4.39WISMNQQDD186 pKa = 4.21RR187 pKa = 11.84VHH189 pKa = 6.92HH190 pKa = 5.97GLQYY194 pKa = 10.78SIQNSNFIQTWTIRR208 pKa = 11.84KK209 pKa = 5.44TAYY212 pKa = 9.0VQFRR216 pKa = 11.84EE217 pKa = 3.82FDD219 pKa = 3.58LVNFPKK225 pKa = 9.35QTT227 pKa = 3.27
MM1 pKa = 7.73PVRR4 pKa = 11.84SRR6 pKa = 11.84YY7 pKa = 7.8SRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WRR14 pKa = 11.84RR15 pKa = 11.84NRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84GPQRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.46ARR28 pKa = 11.84GGYY31 pKa = 8.14RR32 pKa = 11.84WRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 10.0NGIFNARR43 pKa = 11.84LQTEE47 pKa = 4.15EE48 pKa = 4.3TLTVASAATPSWCVGHH64 pKa = 6.73VNVTLKK70 pKa = 11.3SMMPKK75 pKa = 10.66GPGTTTNLYY84 pKa = 10.72GMPFSYY90 pKa = 10.84YY91 pKa = 10.36RR92 pKa = 11.84IRR94 pKa = 11.84KK95 pKa = 8.94LKK97 pKa = 10.45IEE99 pKa = 4.52FLPQIGITSNRR110 pKa = 11.84SYY112 pKa = 11.36GGTAIILDD120 pKa = 3.65GDD122 pKa = 4.31YY123 pKa = 10.79ISEE126 pKa = 4.45GKK128 pKa = 8.66NQTYY132 pKa = 10.76DD133 pKa = 3.31PLANHH138 pKa = 6.51SSRR141 pKa = 11.84HH142 pKa = 4.93NLTNIHH148 pKa = 5.45RR149 pKa = 11.84HH150 pKa = 4.68SRR152 pKa = 11.84YY153 pKa = 7.57FTPKK157 pKa = 8.81PQHH160 pKa = 5.74SSKK163 pKa = 11.04NISTYY168 pKa = 9.15FQPNNKK174 pKa = 8.63QLQEE178 pKa = 4.39WISMNQQDD186 pKa = 4.21RR187 pKa = 11.84VHH189 pKa = 6.92HH190 pKa = 5.97GLQYY194 pKa = 10.78SIQNSNFIQTWTIRR208 pKa = 11.84KK209 pKa = 5.44TAYY212 pKa = 9.0VQFRR216 pKa = 11.84EE217 pKa = 3.82FDD219 pKa = 3.58LVNFPKK225 pKa = 9.35QTT227 pKa = 3.27
Molecular weight: 26.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
524 |
227 |
297 |
262.0 |
30.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.962 ± 1.0 | 1.336 ± 0.623 |
4.198 ± 1.389 | 4.962 ± 1.613 |
3.244 ± 0.501 | 8.397 ± 1.551 |
2.863 ± 0.46 | 4.58 ± 1.104 |
6.298 ± 1.01 | 6.679 ± 0.969 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.099 ± 0.072 | 5.534 ± 1.36 |
4.58 ± 0.491 | 4.771 ± 1.278 |
9.351 ± 2.076 | 4.962 ± 2.065 |
6.87 ± 0.737 | 6.298 ± 1.93 |
2.29 ± 0.061 | 5.725 ± 0.001 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
