
Avian metaavulavirus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
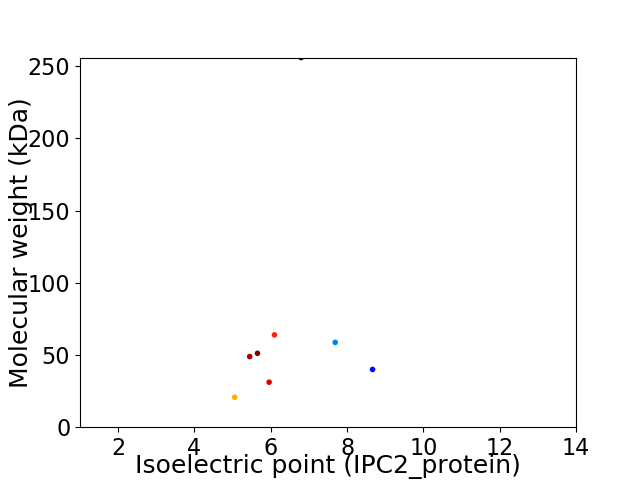
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
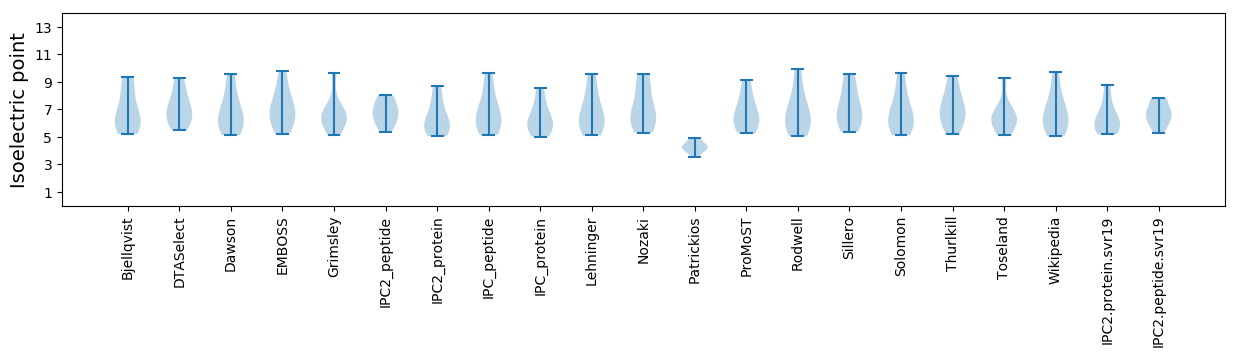
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3X605|D3X605_9MONO Isoform of D3X603 W protein OS=Avian metaavulavirus 5 OX=2560315 GN=P PE=4 SV=1
MM1 pKa = 8.41DD2 pKa = 3.94ITDD5 pKa = 4.13DD6 pKa = 4.33QAILDD11 pKa = 4.9LLTLSNDD18 pKa = 3.38VIEE21 pKa = 5.2SIQHH25 pKa = 5.86AEE27 pKa = 4.29SAGSQPPTYY36 pKa = 10.47GRR38 pKa = 11.84STIPKK43 pKa = 7.35GTTMALTQAWEE54 pKa = 4.24AEE56 pKa = 4.5SNPTQPQPSISPSQLDD72 pKa = 4.19HH73 pKa = 7.13IRR75 pKa = 11.84VDD77 pKa = 3.25QDD79 pKa = 3.6SNKK82 pKa = 9.89NHH84 pKa = 7.36DD85 pKa = 3.78NLKK88 pKa = 9.73NQFDD92 pKa = 4.3SHH94 pKa = 6.21HH95 pKa = 6.46HH96 pKa = 5.82TSQDD100 pKa = 3.6SEE102 pKa = 4.47THH104 pKa = 6.36PNIQVNPSVNRR115 pKa = 11.84TNEE118 pKa = 3.69EE119 pKa = 4.21DD120 pKa = 3.38NSKK123 pKa = 10.42LVQQRR128 pKa = 11.84DD129 pKa = 3.4PSTTWEE135 pKa = 4.53GGSMLDD141 pKa = 4.4RR142 pKa = 11.84EE143 pKa = 4.4LDD145 pKa = 4.25AIQSKK150 pKa = 10.03IKK152 pKa = 10.24RR153 pKa = 11.84GKK155 pKa = 4.42TTKK158 pKa = 10.11SAHH161 pKa = 5.58QPSHH165 pKa = 5.93TATHH169 pKa = 5.9QSPNNLDD176 pKa = 4.28DD177 pKa = 5.61DD178 pKa = 4.3IKK180 pKa = 11.25KK181 pKa = 10.61GGPDD185 pKa = 3.22LRR187 pKa = 6.06
MM1 pKa = 8.41DD2 pKa = 3.94ITDD5 pKa = 4.13DD6 pKa = 4.33QAILDD11 pKa = 4.9LLTLSNDD18 pKa = 3.38VIEE21 pKa = 5.2SIQHH25 pKa = 5.86AEE27 pKa = 4.29SAGSQPPTYY36 pKa = 10.47GRR38 pKa = 11.84STIPKK43 pKa = 7.35GTTMALTQAWEE54 pKa = 4.24AEE56 pKa = 4.5SNPTQPQPSISPSQLDD72 pKa = 4.19HH73 pKa = 7.13IRR75 pKa = 11.84VDD77 pKa = 3.25QDD79 pKa = 3.6SNKK82 pKa = 9.89NHH84 pKa = 7.36DD85 pKa = 3.78NLKK88 pKa = 9.73NQFDD92 pKa = 4.3SHH94 pKa = 6.21HH95 pKa = 6.46HH96 pKa = 5.82TSQDD100 pKa = 3.6SEE102 pKa = 4.47THH104 pKa = 6.36PNIQVNPSVNRR115 pKa = 11.84TNEE118 pKa = 3.69EE119 pKa = 4.21DD120 pKa = 3.38NSKK123 pKa = 10.42LVQQRR128 pKa = 11.84DD129 pKa = 3.4PSTTWEE135 pKa = 4.53GGSMLDD141 pKa = 4.4RR142 pKa = 11.84EE143 pKa = 4.4LDD145 pKa = 4.25AIQSKK150 pKa = 10.03IKK152 pKa = 10.24RR153 pKa = 11.84GKK155 pKa = 4.42TTKK158 pKa = 10.11SAHH161 pKa = 5.58QPSHH165 pKa = 5.93TATHH169 pKa = 5.9QSPNNLDD176 pKa = 4.28DD177 pKa = 5.61DD178 pKa = 4.3IKK180 pKa = 11.25KK181 pKa = 10.61GGPDD185 pKa = 3.22LRR187 pKa = 6.06
Molecular weight: 20.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3X607|D3X607_9MONO Fusion glycoprotein F0 OS=Avian metaavulavirus 5 OX=2560315 GN=F PE=3 SV=1
MM1 pKa = 7.5AQTQLKK7 pKa = 10.42LYY9 pKa = 10.56VNDD12 pKa = 4.07GDD14 pKa = 3.83PSNRR18 pKa = 11.84LLAFPIVMKK27 pKa = 10.67EE28 pKa = 3.8SAQGGKK34 pKa = 9.32VLQPQLRR41 pKa = 11.84ISYY44 pKa = 10.09LGDD47 pKa = 4.42AIGGRR52 pKa = 11.84NSVIFINCYY61 pKa = 10.47GFIEE65 pKa = 4.45SMKK68 pKa = 10.51AGEE71 pKa = 4.93GPFLDD76 pKa = 4.09VNSDD80 pKa = 3.35GKK82 pKa = 11.53GEE84 pKa = 4.29VITAAGLTLGSVTYY98 pKa = 10.66DD99 pKa = 3.2SDD101 pKa = 3.41PTEE104 pKa = 4.21IARR107 pKa = 11.84SCYY110 pKa = 9.83QLLVTVKK117 pKa = 9.94KK118 pKa = 10.79SADD121 pKa = 3.16NTEE124 pKa = 3.97RR125 pKa = 11.84VVYY128 pKa = 8.99TLASKK133 pKa = 10.6PPALSSSRR141 pKa = 11.84VVTSGGCILSAEE153 pKa = 4.41EE154 pKa = 4.56AVKK157 pKa = 10.72CPSKK161 pKa = 10.42LQSGIPYY168 pKa = 9.25KK169 pKa = 10.46FRR171 pKa = 11.84IMFVSLTYY179 pKa = 10.14IHH181 pKa = 6.69QSTLYY186 pKa = 10.21RR187 pKa = 11.84VNNLIAKK194 pKa = 8.94LRR196 pKa = 11.84SPVFISVQLQVTLILDD212 pKa = 4.13LPEE215 pKa = 3.91KK216 pKa = 10.65HH217 pKa = 6.83PMGKK221 pKa = 9.87YY222 pKa = 10.55LINQDD227 pKa = 3.45GVYY230 pKa = 9.88KK231 pKa = 10.47AHH233 pKa = 6.61IWMHH237 pKa = 5.35ICNFKK242 pKa = 9.0KK243 pKa = 9.32TNRR246 pKa = 11.84KK247 pKa = 8.76GADD250 pKa = 2.98RR251 pKa = 11.84SVLQIKK257 pKa = 9.92EE258 pKa = 4.31KK259 pKa = 8.93VRR261 pKa = 11.84KK262 pKa = 8.71MGLKK266 pKa = 8.67VTLADD271 pKa = 3.26LWGPTVVVEE280 pKa = 4.44ATGTMSKK287 pKa = 10.33YY288 pKa = 10.41AVGFFSEE295 pKa = 4.88TKK297 pKa = 10.18VSCHH301 pKa = 6.76PISKK305 pKa = 9.91ISPEE309 pKa = 3.8VAKK312 pKa = 10.5IIWACTTTIGQAVVIIQASSRR333 pKa = 11.84SEE335 pKa = 3.92LLTAEE340 pKa = 5.27DD341 pKa = 4.56IEE343 pKa = 5.03CKK345 pKa = 9.86GTTSIKK351 pKa = 10.21KK352 pKa = 9.33SAVKK356 pKa = 10.21EE357 pKa = 3.82FSLFSKK363 pKa = 9.75PAKK366 pKa = 10.18
MM1 pKa = 7.5AQTQLKK7 pKa = 10.42LYY9 pKa = 10.56VNDD12 pKa = 4.07GDD14 pKa = 3.83PSNRR18 pKa = 11.84LLAFPIVMKK27 pKa = 10.67EE28 pKa = 3.8SAQGGKK34 pKa = 9.32VLQPQLRR41 pKa = 11.84ISYY44 pKa = 10.09LGDD47 pKa = 4.42AIGGRR52 pKa = 11.84NSVIFINCYY61 pKa = 10.47GFIEE65 pKa = 4.45SMKK68 pKa = 10.51AGEE71 pKa = 4.93GPFLDD76 pKa = 4.09VNSDD80 pKa = 3.35GKK82 pKa = 11.53GEE84 pKa = 4.29VITAAGLTLGSVTYY98 pKa = 10.66DD99 pKa = 3.2SDD101 pKa = 3.41PTEE104 pKa = 4.21IARR107 pKa = 11.84SCYY110 pKa = 9.83QLLVTVKK117 pKa = 9.94KK118 pKa = 10.79SADD121 pKa = 3.16NTEE124 pKa = 3.97RR125 pKa = 11.84VVYY128 pKa = 8.99TLASKK133 pKa = 10.6PPALSSSRR141 pKa = 11.84VVTSGGCILSAEE153 pKa = 4.41EE154 pKa = 4.56AVKK157 pKa = 10.72CPSKK161 pKa = 10.42LQSGIPYY168 pKa = 9.25KK169 pKa = 10.46FRR171 pKa = 11.84IMFVSLTYY179 pKa = 10.14IHH181 pKa = 6.69QSTLYY186 pKa = 10.21RR187 pKa = 11.84VNNLIAKK194 pKa = 8.94LRR196 pKa = 11.84SPVFISVQLQVTLILDD212 pKa = 4.13LPEE215 pKa = 3.91KK216 pKa = 10.65HH217 pKa = 6.83PMGKK221 pKa = 9.87YY222 pKa = 10.55LINQDD227 pKa = 3.45GVYY230 pKa = 9.88KK231 pKa = 10.47AHH233 pKa = 6.61IWMHH237 pKa = 5.35ICNFKK242 pKa = 9.0KK243 pKa = 9.32TNRR246 pKa = 11.84KK247 pKa = 8.76GADD250 pKa = 2.98RR251 pKa = 11.84SVLQIKK257 pKa = 9.92EE258 pKa = 4.31KK259 pKa = 8.93VRR261 pKa = 11.84KK262 pKa = 8.71MGLKK266 pKa = 8.67VTLADD271 pKa = 3.26LWGPTVVVEE280 pKa = 4.44ATGTMSKK287 pKa = 10.33YY288 pKa = 10.41AVGFFSEE295 pKa = 4.88TKK297 pKa = 10.18VSCHH301 pKa = 6.76PISKK305 pKa = 9.91ISPEE309 pKa = 3.8VAKK312 pKa = 10.5IIWACTTTIGQAVVIIQASSRR333 pKa = 11.84SEE335 pKa = 3.92LLTAEE340 pKa = 5.27DD341 pKa = 4.56IEE343 pKa = 5.03CKK345 pKa = 9.86GTTSIKK351 pKa = 10.21KK352 pKa = 9.33SAVKK356 pKa = 10.21EE357 pKa = 3.82FSLFSKK363 pKa = 9.75PAKK366 pKa = 10.18
Molecular weight: 40.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5122 |
187 |
2263 |
640.3 |
71.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.111 ± 0.723 | 2.148 ± 0.337 |
4.979 ± 0.695 | 4.666 ± 0.249 |
3.182 ± 0.498 | 5.037 ± 0.361 |
2.382 ± 0.393 | 7.927 ± 0.598 |
4.861 ± 0.534 | 10.406 ± 0.996 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.245 ± 0.288 | 5.174 ± 0.637 |
4.861 ± 0.539 | 5.271 ± 0.575 |
4.627 ± 0.319 | 9.762 ± 0.428 |
7.321 ± 0.432 | 5.018 ± 0.672 |
1.132 ± 0.235 | 2.889 ± 0.528 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
