
Paraburkholderia ribeironis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
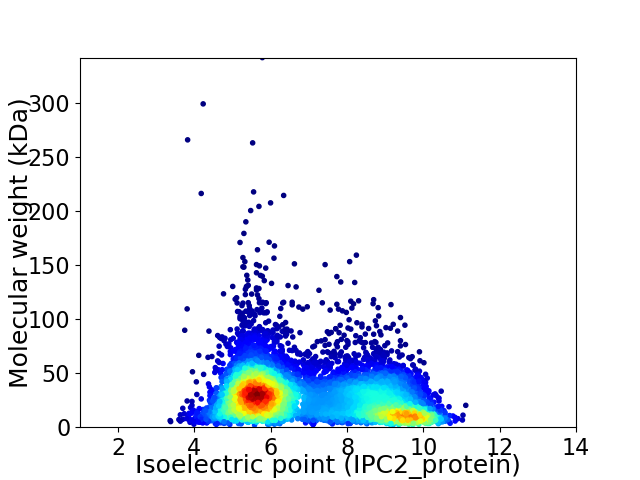
Virtual 2D-PAGE plot for 7335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N7RLZ1|A0A1N7RLZ1_9BURK KAP NTPase domain-containing protein OS=Paraburkholderia ribeironis OX=1247936 GN=BN2475_70104 PE=4 SV=1
MM1 pKa = 7.22RR2 pKa = 11.84HH3 pKa = 5.85LRR5 pKa = 11.84HH6 pKa = 6.55EE7 pKa = 3.92YY8 pKa = 10.22RR9 pKa = 11.84IRR11 pKa = 11.84LVQRR15 pKa = 11.84SASKK19 pKa = 10.4DD20 pKa = 3.64AAWFAAGKK28 pKa = 8.13PSKK31 pKa = 8.87VTRR34 pKa = 11.84HH35 pKa = 5.54PLWMRR40 pKa = 11.84SVAALMATVMYY51 pKa = 7.68FAPAVFLADD60 pKa = 3.3ATAYY64 pKa = 10.36AAVIVDD70 pKa = 3.67PRR72 pKa = 11.84APVPFQPTVTQGSTGAGIVNITAPNAQGLSVNQFQSLSVGAPGLVFNNSRR122 pKa = 11.84MAGTSLIGGPVGANPNLVGRR142 pKa = 11.84TASMILAQVTSTGAQYY158 pKa = 10.95RR159 pKa = 11.84SVLAGPVEE167 pKa = 4.57VFGAPASLIIANPNGVSIPGGMQVTNASSLTLTTGVPQFVTGPGGVTTDD216 pKa = 3.58YY217 pKa = 11.53AGAGALAYY225 pKa = 9.79DD226 pKa = 4.14VRR228 pKa = 11.84SGDD231 pKa = 3.25IAINGAPGNDD241 pKa = 3.7GTPGPGIDD249 pKa = 3.51GTVGNLDD256 pKa = 5.54LIAQTLSISAPLKK269 pKa = 9.86ADD271 pKa = 3.15QRR273 pKa = 11.84VNLIAGNQLVSPTATGAAGTSYY295 pKa = 9.3GTASNGAANNAAVIQTPGGLAIDD318 pKa = 4.38ASQYY322 pKa = 11.18GSVTSGQIYY331 pKa = 10.05IVSTAAGMGVNTQGALAATAGNVTVNTNGDD361 pKa = 3.0ITAGNTFANQNVTLTSAGTTNVFGTGLANQNFTINANGDD400 pKa = 3.29INATGPVSAGQNVSLNAGGDD420 pKa = 3.66LNAASVAANGSADD433 pKa = 4.6LIAGGSMSLGSLSAHH448 pKa = 6.79DD449 pKa = 5.11LDD451 pKa = 5.59IEE453 pKa = 4.3ATNGNLTLGQAITAPGTIRR472 pKa = 11.84AVAGLDD478 pKa = 3.28LTVNGSVQGGGTVGLSATRR497 pKa = 11.84NAAVNGTVTGVGDD510 pKa = 3.66TSVTGITGTSNISGNVQTNGALTVSSAQGTTLGGIVQAQGPVSVIAQNGSITGQGDD566 pKa = 3.57VASSQGSVSLDD577 pKa = 2.8AGQAINLSGSVQVGGSITALAGGDD601 pKa = 3.53ALFGGTLAAPGAISVTTGGDD621 pKa = 3.02ATLGGNATSGSTLTIAAGANASILGTAASVGNMSLTAANGSLSTANNVVTLGTLAASGQQGVNLGGNVYY690 pKa = 10.51SQGNAQISSGAGSVAVAGALSTPGSITVNAAQDD723 pKa = 3.61VTVTGLLNSGGDD735 pKa = 3.55ATITATRR742 pKa = 11.84DD743 pKa = 3.01ANLNVGLDD751 pKa = 3.62VQGTGNATITVGRR764 pKa = 11.84DD765 pKa = 3.05INGSGAVNVANDD777 pKa = 3.73TTLLAGSNIGISGAIQTGNNLAVTAGNNLAVGATTAVGNEE817 pKa = 4.19TLTATAGSATLASDD831 pKa = 3.81ALSGGDD837 pKa = 3.36MAIRR841 pKa = 11.84AAIDD845 pKa = 3.23VTTPGSVQSLGNLNVNAQGGNLSASGRR872 pKa = 11.84VSTAGTATLNAGQNLSLAGQTIVSGDD898 pKa = 3.41ATLTGANITTQGLAVGGNLTATAQNSLDD926 pKa = 3.57TSAGQLNAAFDD937 pKa = 3.98ASAPALSVNGNATLTGANVTTANAVIGGIYY967 pKa = 9.86SATGTTSIMTGGTAAYY983 pKa = 9.72QSNATLAGGTVTNVGTQMAAGDD1005 pKa = 4.06LTVSGSTVTNQGALSSLQTAAVNATDD1031 pKa = 4.7LNNSGSIYY1039 pKa = 9.43GTAANLNVTNSTVNAGALLATNTLALTTASLDD1071 pKa = 3.57NSNGLIFAGDD1081 pKa = 3.82VNSPTSPVGDD1091 pKa = 3.17VTVNVNGGNGTFNNTNGQILAQNGLTLNLPNQVIDD1126 pKa = 4.12PSAATMGMLNGGSAFNLSAQSISNTGAWTLPGTAVTVIAAQGMSNTGTINQGAGTLALNGAVSNAGTIAAQDD1198 pKa = 3.65LTITGSLANQTGALVQANDD1217 pKa = 3.39AFTLNGSGTNMGTVQAVNSLTIAGSGYY1244 pKa = 10.73DD1245 pKa = 3.5NSNGITQAGNSSSAAGSGNVNISLSGDD1272 pKa = 3.4LTNAGGMLTATNDD1285 pKa = 3.91LSITANNVVNSGAGADD1301 pKa = 3.75PGVTTTTTTTVDD1313 pKa = 3.38NPGLALALVVGSDD1326 pKa = 4.63KK1327 pKa = 10.53IDD1329 pKa = 3.76EE1330 pKa = 4.37AATWGSGGVKK1340 pKa = 9.7FCCTVYY1346 pKa = 11.08GLGTNQVTLADD1357 pKa = 4.14ALSVDD1362 pKa = 4.29GVADD1366 pKa = 3.53PTSINTILGPTAAAWKK1382 pKa = 9.88QFSPPVTNGNTVTFVEE1398 pKa = 4.82IPTVTGTSNTGNSIIQNLWFVQTPDD1423 pKa = 3.33NASQAIATITVVLPTATEE1441 pKa = 4.24TTTTTGGTAGGTSVIAAGHH1460 pKa = 5.51NVSINASSLNNQGGKK1475 pKa = 9.86VSAGNDD1481 pKa = 3.15TSVNVQSLNNGGSVYY1496 pKa = 10.36TSTVTDD1502 pKa = 3.79TVDD1505 pKa = 3.02VASIDD1510 pKa = 3.83SFLSQAPSTISLWNTFNGPNGLEE1533 pKa = 4.06GCPFGMAGVCIDD1545 pKa = 3.67PSGIRR1550 pKa = 11.84LAAPGTVTPLSTSSSITVQGAAGQIVAGHH1579 pKa = 7.04DD1580 pKa = 3.83LNLSGGDD1587 pKa = 3.58LVNAGTLAATNDD1599 pKa = 3.76VNISAASFTNQGTSTGTMTTTAGCAAGFSAGCTTLSTTNPNAQTYY1644 pKa = 10.11SYY1646 pKa = 10.64QQINSNVTAGHH1657 pKa = 7.25DD1658 pKa = 3.25IVIAANTVSNTYY1670 pKa = 11.09GNLAAGNSVVIGGAGTTAAAPTQAASVTNTSGAIAAGNDD1709 pKa = 2.93VDD1711 pKa = 5.02INGATLVNTIAAPVQVHH1728 pKa = 5.19QNYY1731 pKa = 7.67GTATPFTGCTSNCEE1745 pKa = 3.93AYY1747 pKa = 9.71IDD1749 pKa = 4.09VQSATPATITANHH1762 pKa = 6.17NVNLTAGSFSNTGSLITALNNVTINATATAGSDD1795 pKa = 3.37NQYY1798 pKa = 11.49LNAYY1802 pKa = 6.39WASDD1806 pKa = 3.46LLHH1809 pKa = 6.7YY1810 pKa = 7.46NTSYY1814 pKa = 10.76VAWGCASNPSLCQTLYY1830 pKa = 11.02GNAYY1834 pKa = 9.78SSSAAQDD1841 pKa = 3.63PAGLPSSVGLPDD1853 pKa = 4.28FVPATIQAGNTLSVNSPTLTNTGNVVGQTVALSGSQLVNGLMDD1896 pKa = 5.37PDD1898 pKa = 4.9VYY1900 pKa = 10.79TPPPVVTGQVISLGPPAVPANVATTVNGSWQVTNANGQAVSVTGSAGLPSNSPIGVQTVGKK1961 pKa = 8.64PLAPLVSQAGAPAGSTVQTIAGQTIPVTYY1990 pKa = 10.0LVNSPASAVTGDD2002 pKa = 4.53LSPAALLAALPANLQPGTTQFYY2024 pKa = 10.09YY2025 pKa = 10.74DD2026 pKa = 5.26PYY2028 pKa = 9.32TQAQQVEE2035 pKa = 4.56QAALQATGKK2044 pKa = 10.51ASFYY2048 pKa = 9.49STTGATDD2055 pKa = 3.2STGQASISNQDD2066 pKa = 3.0TQALYY2071 pKa = 10.66GAALRR2076 pKa = 11.84YY2077 pKa = 9.93AEE2079 pKa = 4.26QNNVALGTQLSAAQLAQINAPMLWYY2104 pKa = 10.39VEE2106 pKa = 4.05QSVPEE2111 pKa = 4.31PGCTVTGSGACPTVQALMPEE2131 pKa = 4.14VLLPQNYY2138 pKa = 10.12AEE2140 pKa = 4.23VSADD2144 pKa = 3.7GEE2146 pKa = 4.08ISGANVALNYY2156 pKa = 10.7ANSILNTGSISAQNLTVNTASLTNEE2181 pKa = 3.73EE2182 pKa = 4.38RR2183 pKa = 11.84STNIGTIYY2191 pKa = 10.78NNVDD2195 pKa = 2.84NGVAVTTGTLVQQGGFMSAMNYY2217 pKa = 10.24DD2218 pKa = 4.6LNAQTIDD2225 pKa = 3.62QIGGALQQINSDD2237 pKa = 3.41GSVNQAATSQMLANLKK2253 pKa = 9.98SQLGSNFTQSTVSNNLNTTVISDD2276 pKa = 3.39GGMGLMTVFEE2286 pKa = 4.25MAIIVAMSVVTAGAASAALAGMQVAAADD2314 pKa = 3.59AGMATLAAGGTIVEE2328 pKa = 4.45ANAAAGILASSAFAAGGVANAAIAGAMAGLVGSAAGDD2365 pKa = 3.7LMSSGTLNFGDD2376 pKa = 4.08VLKK2379 pKa = 10.84GAAIGGLTAGLTDD2392 pKa = 6.4GITLDD2397 pKa = 3.79DD2398 pKa = 3.98SGIGFSLSGSPDD2410 pKa = 3.17SLASLAGVQSVGNSLVPQAGASTAGSLPEE2439 pKa = 3.95QALALAGEE2447 pKa = 4.49ATVQAGVQTAIDD2459 pKa = 4.03GGSFLASLRR2468 pKa = 11.84NSAVSDD2474 pKa = 3.63VAAAGAYY2481 pKa = 10.24AIGNANLEE2489 pKa = 4.28GDD2491 pKa = 4.45FGTGTQQEE2499 pKa = 4.57LEE2501 pKa = 4.44YY2502 pKa = 10.54VAAHH2506 pKa = 6.67AALGCAASAAEE2517 pKa = 4.25GTGCVGGAIGGAVSAALSPSFIGLIDD2543 pKa = 3.84PVANPLDD2550 pKa = 4.22SGQLAALAAFATLAGGGLAALAGVNVQGAATAAQNEE2586 pKa = 4.31ALNNSSVHH2594 pKa = 6.87IGDD2597 pKa = 4.74GKK2599 pKa = 11.16SEE2601 pKa = 4.2TQKK2604 pKa = 11.06EE2605 pKa = 4.09LDD2607 pKa = 3.63EE2608 pKa = 5.77LRR2610 pKa = 11.84AQQAKK2615 pKa = 9.73EE2616 pKa = 3.65KK2617 pKa = 10.3AALSEE2622 pKa = 4.14ATGGQVTDD2630 pKa = 3.92NSAATVPGSSLTAGLGGATNNAPLGLGSTGRR2661 pKa = 11.84AVSNNLQEE2669 pKa = 4.68QLALQQAMSNPAAGVQLPIPLGDD2692 pKa = 4.1SRR2694 pKa = 11.84WPAADD2699 pKa = 2.99GWVKK2703 pKa = 10.62MSQNVNGVEE2712 pKa = 3.49IHH2714 pKa = 5.49YY2715 pKa = 10.01VRR2717 pKa = 11.84NVNSGAVDD2725 pKa = 3.57DD2726 pKa = 5.43FKK2728 pKa = 11.64FKK2730 pKa = 11.12
MM1 pKa = 7.22RR2 pKa = 11.84HH3 pKa = 5.85LRR5 pKa = 11.84HH6 pKa = 6.55EE7 pKa = 3.92YY8 pKa = 10.22RR9 pKa = 11.84IRR11 pKa = 11.84LVQRR15 pKa = 11.84SASKK19 pKa = 10.4DD20 pKa = 3.64AAWFAAGKK28 pKa = 8.13PSKK31 pKa = 8.87VTRR34 pKa = 11.84HH35 pKa = 5.54PLWMRR40 pKa = 11.84SVAALMATVMYY51 pKa = 7.68FAPAVFLADD60 pKa = 3.3ATAYY64 pKa = 10.36AAVIVDD70 pKa = 3.67PRR72 pKa = 11.84APVPFQPTVTQGSTGAGIVNITAPNAQGLSVNQFQSLSVGAPGLVFNNSRR122 pKa = 11.84MAGTSLIGGPVGANPNLVGRR142 pKa = 11.84TASMILAQVTSTGAQYY158 pKa = 10.95RR159 pKa = 11.84SVLAGPVEE167 pKa = 4.57VFGAPASLIIANPNGVSIPGGMQVTNASSLTLTTGVPQFVTGPGGVTTDD216 pKa = 3.58YY217 pKa = 11.53AGAGALAYY225 pKa = 9.79DD226 pKa = 4.14VRR228 pKa = 11.84SGDD231 pKa = 3.25IAINGAPGNDD241 pKa = 3.7GTPGPGIDD249 pKa = 3.51GTVGNLDD256 pKa = 5.54LIAQTLSISAPLKK269 pKa = 9.86ADD271 pKa = 3.15QRR273 pKa = 11.84VNLIAGNQLVSPTATGAAGTSYY295 pKa = 9.3GTASNGAANNAAVIQTPGGLAIDD318 pKa = 4.38ASQYY322 pKa = 11.18GSVTSGQIYY331 pKa = 10.05IVSTAAGMGVNTQGALAATAGNVTVNTNGDD361 pKa = 3.0ITAGNTFANQNVTLTSAGTTNVFGTGLANQNFTINANGDD400 pKa = 3.29INATGPVSAGQNVSLNAGGDD420 pKa = 3.66LNAASVAANGSADD433 pKa = 4.6LIAGGSMSLGSLSAHH448 pKa = 6.79DD449 pKa = 5.11LDD451 pKa = 5.59IEE453 pKa = 4.3ATNGNLTLGQAITAPGTIRR472 pKa = 11.84AVAGLDD478 pKa = 3.28LTVNGSVQGGGTVGLSATRR497 pKa = 11.84NAAVNGTVTGVGDD510 pKa = 3.66TSVTGITGTSNISGNVQTNGALTVSSAQGTTLGGIVQAQGPVSVIAQNGSITGQGDD566 pKa = 3.57VASSQGSVSLDD577 pKa = 2.8AGQAINLSGSVQVGGSITALAGGDD601 pKa = 3.53ALFGGTLAAPGAISVTTGGDD621 pKa = 3.02ATLGGNATSGSTLTIAAGANASILGTAASVGNMSLTAANGSLSTANNVVTLGTLAASGQQGVNLGGNVYY690 pKa = 10.51SQGNAQISSGAGSVAVAGALSTPGSITVNAAQDD723 pKa = 3.61VTVTGLLNSGGDD735 pKa = 3.55ATITATRR742 pKa = 11.84DD743 pKa = 3.01ANLNVGLDD751 pKa = 3.62VQGTGNATITVGRR764 pKa = 11.84DD765 pKa = 3.05INGSGAVNVANDD777 pKa = 3.73TTLLAGSNIGISGAIQTGNNLAVTAGNNLAVGATTAVGNEE817 pKa = 4.19TLTATAGSATLASDD831 pKa = 3.81ALSGGDD837 pKa = 3.36MAIRR841 pKa = 11.84AAIDD845 pKa = 3.23VTTPGSVQSLGNLNVNAQGGNLSASGRR872 pKa = 11.84VSTAGTATLNAGQNLSLAGQTIVSGDD898 pKa = 3.41ATLTGANITTQGLAVGGNLTATAQNSLDD926 pKa = 3.57TSAGQLNAAFDD937 pKa = 3.98ASAPALSVNGNATLTGANVTTANAVIGGIYY967 pKa = 9.86SATGTTSIMTGGTAAYY983 pKa = 9.72QSNATLAGGTVTNVGTQMAAGDD1005 pKa = 4.06LTVSGSTVTNQGALSSLQTAAVNATDD1031 pKa = 4.7LNNSGSIYY1039 pKa = 9.43GTAANLNVTNSTVNAGALLATNTLALTTASLDD1071 pKa = 3.57NSNGLIFAGDD1081 pKa = 3.82VNSPTSPVGDD1091 pKa = 3.17VTVNVNGGNGTFNNTNGQILAQNGLTLNLPNQVIDD1126 pKa = 4.12PSAATMGMLNGGSAFNLSAQSISNTGAWTLPGTAVTVIAAQGMSNTGTINQGAGTLALNGAVSNAGTIAAQDD1198 pKa = 3.65LTITGSLANQTGALVQANDD1217 pKa = 3.39AFTLNGSGTNMGTVQAVNSLTIAGSGYY1244 pKa = 10.73DD1245 pKa = 3.5NSNGITQAGNSSSAAGSGNVNISLSGDD1272 pKa = 3.4LTNAGGMLTATNDD1285 pKa = 3.91LSITANNVVNSGAGADD1301 pKa = 3.75PGVTTTTTTTVDD1313 pKa = 3.38NPGLALALVVGSDD1326 pKa = 4.63KK1327 pKa = 10.53IDD1329 pKa = 3.76EE1330 pKa = 4.37AATWGSGGVKK1340 pKa = 9.7FCCTVYY1346 pKa = 11.08GLGTNQVTLADD1357 pKa = 4.14ALSVDD1362 pKa = 4.29GVADD1366 pKa = 3.53PTSINTILGPTAAAWKK1382 pKa = 9.88QFSPPVTNGNTVTFVEE1398 pKa = 4.82IPTVTGTSNTGNSIIQNLWFVQTPDD1423 pKa = 3.33NASQAIATITVVLPTATEE1441 pKa = 4.24TTTTTGGTAGGTSVIAAGHH1460 pKa = 5.51NVSINASSLNNQGGKK1475 pKa = 9.86VSAGNDD1481 pKa = 3.15TSVNVQSLNNGGSVYY1496 pKa = 10.36TSTVTDD1502 pKa = 3.79TVDD1505 pKa = 3.02VASIDD1510 pKa = 3.83SFLSQAPSTISLWNTFNGPNGLEE1533 pKa = 4.06GCPFGMAGVCIDD1545 pKa = 3.67PSGIRR1550 pKa = 11.84LAAPGTVTPLSTSSSITVQGAAGQIVAGHH1579 pKa = 7.04DD1580 pKa = 3.83LNLSGGDD1587 pKa = 3.58LVNAGTLAATNDD1599 pKa = 3.76VNISAASFTNQGTSTGTMTTTAGCAAGFSAGCTTLSTTNPNAQTYY1644 pKa = 10.11SYY1646 pKa = 10.64QQINSNVTAGHH1657 pKa = 7.25DD1658 pKa = 3.25IVIAANTVSNTYY1670 pKa = 11.09GNLAAGNSVVIGGAGTTAAAPTQAASVTNTSGAIAAGNDD1709 pKa = 2.93VDD1711 pKa = 5.02INGATLVNTIAAPVQVHH1728 pKa = 5.19QNYY1731 pKa = 7.67GTATPFTGCTSNCEE1745 pKa = 3.93AYY1747 pKa = 9.71IDD1749 pKa = 4.09VQSATPATITANHH1762 pKa = 6.17NVNLTAGSFSNTGSLITALNNVTINATATAGSDD1795 pKa = 3.37NQYY1798 pKa = 11.49LNAYY1802 pKa = 6.39WASDD1806 pKa = 3.46LLHH1809 pKa = 6.7YY1810 pKa = 7.46NTSYY1814 pKa = 10.76VAWGCASNPSLCQTLYY1830 pKa = 11.02GNAYY1834 pKa = 9.78SSSAAQDD1841 pKa = 3.63PAGLPSSVGLPDD1853 pKa = 4.28FVPATIQAGNTLSVNSPTLTNTGNVVGQTVALSGSQLVNGLMDD1896 pKa = 5.37PDD1898 pKa = 4.9VYY1900 pKa = 10.79TPPPVVTGQVISLGPPAVPANVATTVNGSWQVTNANGQAVSVTGSAGLPSNSPIGVQTVGKK1961 pKa = 8.64PLAPLVSQAGAPAGSTVQTIAGQTIPVTYY1990 pKa = 10.0LVNSPASAVTGDD2002 pKa = 4.53LSPAALLAALPANLQPGTTQFYY2024 pKa = 10.09YY2025 pKa = 10.74DD2026 pKa = 5.26PYY2028 pKa = 9.32TQAQQVEE2035 pKa = 4.56QAALQATGKK2044 pKa = 10.51ASFYY2048 pKa = 9.49STTGATDD2055 pKa = 3.2STGQASISNQDD2066 pKa = 3.0TQALYY2071 pKa = 10.66GAALRR2076 pKa = 11.84YY2077 pKa = 9.93AEE2079 pKa = 4.26QNNVALGTQLSAAQLAQINAPMLWYY2104 pKa = 10.39VEE2106 pKa = 4.05QSVPEE2111 pKa = 4.31PGCTVTGSGACPTVQALMPEE2131 pKa = 4.14VLLPQNYY2138 pKa = 10.12AEE2140 pKa = 4.23VSADD2144 pKa = 3.7GEE2146 pKa = 4.08ISGANVALNYY2156 pKa = 10.7ANSILNTGSISAQNLTVNTASLTNEE2181 pKa = 3.73EE2182 pKa = 4.38RR2183 pKa = 11.84STNIGTIYY2191 pKa = 10.78NNVDD2195 pKa = 2.84NGVAVTTGTLVQQGGFMSAMNYY2217 pKa = 10.24DD2218 pKa = 4.6LNAQTIDD2225 pKa = 3.62QIGGALQQINSDD2237 pKa = 3.41GSVNQAATSQMLANLKK2253 pKa = 9.98SQLGSNFTQSTVSNNLNTTVISDD2276 pKa = 3.39GGMGLMTVFEE2286 pKa = 4.25MAIIVAMSVVTAGAASAALAGMQVAAADD2314 pKa = 3.59AGMATLAAGGTIVEE2328 pKa = 4.45ANAAAGILASSAFAAGGVANAAIAGAMAGLVGSAAGDD2365 pKa = 3.7LMSSGTLNFGDD2376 pKa = 4.08VLKK2379 pKa = 10.84GAAIGGLTAGLTDD2392 pKa = 6.4GITLDD2397 pKa = 3.79DD2398 pKa = 3.98SGIGFSLSGSPDD2410 pKa = 3.17SLASLAGVQSVGNSLVPQAGASTAGSLPEE2439 pKa = 3.95QALALAGEE2447 pKa = 4.49ATVQAGVQTAIDD2459 pKa = 4.03GGSFLASLRR2468 pKa = 11.84NSAVSDD2474 pKa = 3.63VAAAGAYY2481 pKa = 10.24AIGNANLEE2489 pKa = 4.28GDD2491 pKa = 4.45FGTGTQQEE2499 pKa = 4.57LEE2501 pKa = 4.44YY2502 pKa = 10.54VAAHH2506 pKa = 6.67AALGCAASAAEE2517 pKa = 4.25GTGCVGGAIGGAVSAALSPSFIGLIDD2543 pKa = 3.84PVANPLDD2550 pKa = 4.22SGQLAALAAFATLAGGGLAALAGVNVQGAATAAQNEE2586 pKa = 4.31ALNNSSVHH2594 pKa = 6.87IGDD2597 pKa = 4.74GKK2599 pKa = 11.16SEE2601 pKa = 4.2TQKK2604 pKa = 11.06EE2605 pKa = 4.09LDD2607 pKa = 3.63EE2608 pKa = 5.77LRR2610 pKa = 11.84AQQAKK2615 pKa = 9.73EE2616 pKa = 3.65KK2617 pKa = 10.3AALSEE2622 pKa = 4.14ATGGQVTDD2630 pKa = 3.92NSAATVPGSSLTAGLGGATNNAPLGLGSTGRR2661 pKa = 11.84AVSNNLQEE2669 pKa = 4.68QLALQQAMSNPAAGVQLPIPLGDD2692 pKa = 4.1SRR2694 pKa = 11.84WPAADD2699 pKa = 2.99GWVKK2703 pKa = 10.62MSQNVNGVEE2712 pKa = 3.49IHH2714 pKa = 5.49YY2715 pKa = 10.01VRR2717 pKa = 11.84NVNSGAVDD2725 pKa = 3.57DD2726 pKa = 5.43FKK2728 pKa = 11.64FKK2730 pKa = 11.12
Molecular weight: 265.78 kDa
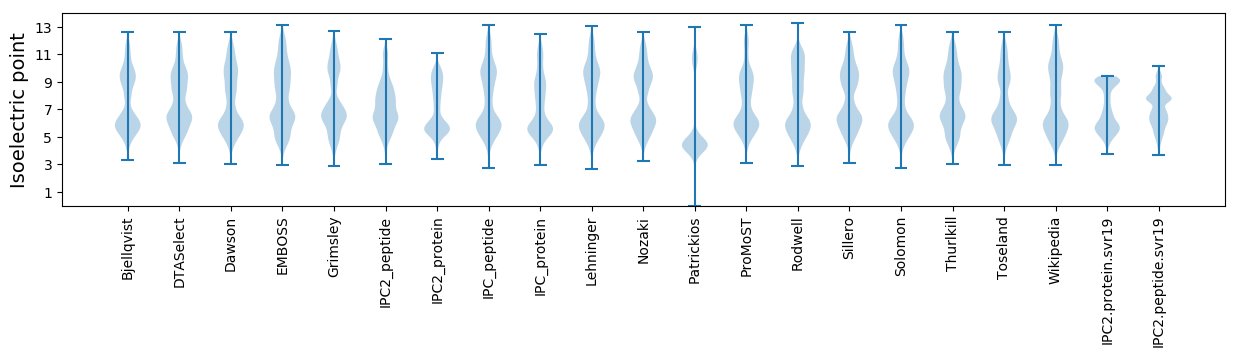
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7SF25|A0A1N7SF25_9BURK Uncharacterized protein OS=Paraburkholderia ribeironis OX=1247936 GN=BN2475_570059 PE=4 SV=1
MM1 pKa = 7.64LLPRR5 pKa = 11.84RR6 pKa = 11.84LGRR9 pKa = 11.84LGLLGLRR16 pKa = 11.84RR17 pKa = 11.84PLGRR21 pKa = 11.84RR22 pKa = 11.84SQRR25 pKa = 11.84LSKK28 pKa = 10.7APRR31 pKa = 11.84RR32 pKa = 11.84PLMGSALYY40 pKa = 9.26EE41 pKa = 3.77RR42 pKa = 11.84SMGTRR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 10.13RR50 pKa = 11.84PTRR53 pKa = 11.84VNSRR57 pKa = 3.22
MM1 pKa = 7.64LLPRR5 pKa = 11.84RR6 pKa = 11.84LGRR9 pKa = 11.84LGLLGLRR16 pKa = 11.84RR17 pKa = 11.84PLGRR21 pKa = 11.84RR22 pKa = 11.84SQRR25 pKa = 11.84LSKK28 pKa = 10.7APRR31 pKa = 11.84RR32 pKa = 11.84PLMGSALYY40 pKa = 9.26EE41 pKa = 3.77RR42 pKa = 11.84SMGTRR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 10.13RR50 pKa = 11.84PTRR53 pKa = 11.84VNSRR57 pKa = 3.22
Molecular weight: 6.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2089330 |
20 |
3182 |
284.8 |
30.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.57 ± 0.048 | 1.068 ± 0.012 |
5.376 ± 0.021 | 5.042 ± 0.032 |
3.678 ± 0.016 | 8.001 ± 0.027 |
2.43 ± 0.013 | 4.71 ± 0.021 |
3.108 ± 0.024 | 10.095 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.384 ± 0.015 | 2.89 ± 0.02 |
5.098 ± 0.022 | 3.725 ± 0.021 |
7.183 ± 0.036 | 5.829 ± 0.028 |
5.425 ± 0.024 | 7.607 ± 0.026 |
1.374 ± 0.013 | 2.406 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
