
Frangipani mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
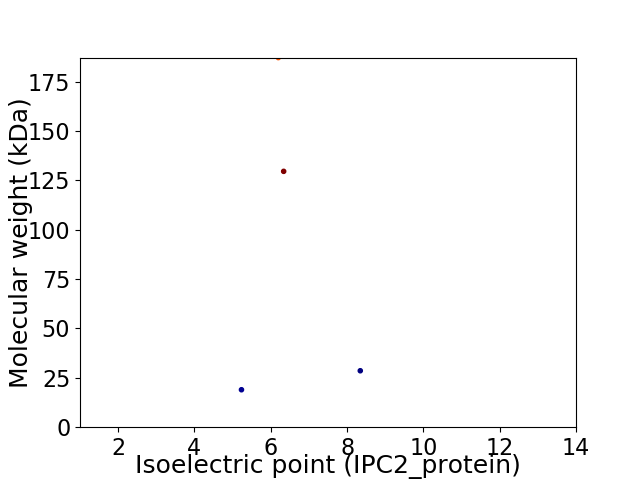
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2F1P9|E2F1P9_9VIRU Capsid protein OS=Frangipani mosaic virus OX=99585 PE=3 SV=1
MM1 pKa = 7.89SYY3 pKa = 11.05TNISTTNIIYY13 pKa = 10.22LARR16 pKa = 11.84SYY18 pKa = 11.08IPGVDD23 pKa = 4.58FIDD26 pKa = 5.69LIISGLGQSFQTQSARR42 pKa = 11.84DD43 pKa = 3.59EE44 pKa = 4.37FRR46 pKa = 11.84SQLIGSYY53 pKa = 10.02KK54 pKa = 9.83VVVSKK59 pKa = 7.95TVRR62 pKa = 11.84FPEE65 pKa = 3.67NTIYY69 pKa = 10.89LWANNPTIRR78 pKa = 11.84PLLHH82 pKa = 7.05AVFQALDD89 pKa = 3.32TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.13VEE98 pKa = 4.19STNVVNPTSSEE109 pKa = 3.78TRR111 pKa = 11.84EE112 pKa = 3.72ATRR115 pKa = 11.84RR116 pKa = 11.84VDD118 pKa = 3.47DD119 pKa = 3.42ATVAVRR125 pKa = 11.84SQLQLLFDD133 pKa = 4.5ALSGGSGLYY142 pKa = 9.68DD143 pKa = 3.54RR144 pKa = 11.84KK145 pKa = 10.73AFEE148 pKa = 4.72DD149 pKa = 3.62ASGLVWEE156 pKa = 4.79EE157 pKa = 3.51AAAVGTAGTSGTGTTTAA174 pKa = 4.32
MM1 pKa = 7.89SYY3 pKa = 11.05TNISTTNIIYY13 pKa = 10.22LARR16 pKa = 11.84SYY18 pKa = 11.08IPGVDD23 pKa = 4.58FIDD26 pKa = 5.69LIISGLGQSFQTQSARR42 pKa = 11.84DD43 pKa = 3.59EE44 pKa = 4.37FRR46 pKa = 11.84SQLIGSYY53 pKa = 10.02KK54 pKa = 9.83VVVSKK59 pKa = 7.95TVRR62 pKa = 11.84FPEE65 pKa = 3.67NTIYY69 pKa = 10.89LWANNPTIRR78 pKa = 11.84PLLHH82 pKa = 7.05AVFQALDD89 pKa = 3.32TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.13VEE98 pKa = 4.19STNVVNPTSSEE109 pKa = 3.78TRR111 pKa = 11.84EE112 pKa = 3.72ATRR115 pKa = 11.84RR116 pKa = 11.84VDD118 pKa = 3.47DD119 pKa = 3.42ATVAVRR125 pKa = 11.84SQLQLLFDD133 pKa = 4.5ALSGGSGLYY142 pKa = 9.68DD143 pKa = 3.54RR144 pKa = 11.84KK145 pKa = 10.73AFEE148 pKa = 4.72DD149 pKa = 3.62ASGLVWEE156 pKa = 4.79EE157 pKa = 3.51AAAVGTAGTSGTGTTTAA174 pKa = 4.32
Molecular weight: 18.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2F1P9|E2F1P9_9VIRU Capsid protein OS=Frangipani mosaic virus OX=99585 PE=3 SV=1
MM1 pKa = 7.48ALVEE5 pKa = 4.33LKK7 pKa = 10.38EE8 pKa = 4.08PKK10 pKa = 9.52QLKK13 pKa = 10.49VNDD16 pKa = 3.94FVKK19 pKa = 10.43MSFADD24 pKa = 3.98KK25 pKa = 10.36ILPRR29 pKa = 11.84SLTRR33 pKa = 11.84LRR35 pKa = 11.84TVSISEE41 pKa = 4.26TNVVKK46 pKa = 10.9LSGLGSTVNLNILKK60 pKa = 10.4GVVLNSEE67 pKa = 4.39SKK69 pKa = 9.31YY70 pKa = 7.4VTIRR74 pKa = 11.84GVVISGVWMVPEE86 pKa = 4.63GCGGGATVTLMDD98 pKa = 4.15RR99 pKa = 11.84RR100 pKa = 11.84MKK102 pKa = 10.6GFKK105 pKa = 9.92NGLVAEE111 pKa = 5.18FKK113 pKa = 10.21TRR115 pKa = 11.84ASSRR119 pKa = 11.84DD120 pKa = 3.52FQFKK124 pKa = 10.15FIPNYY129 pKa = 10.04SMCVDD134 pKa = 3.75DD135 pKa = 5.38VKK137 pKa = 10.74RR138 pKa = 11.84APWEE142 pKa = 3.88LFFKK146 pKa = 10.93LVGVPIEE153 pKa = 4.42DD154 pKa = 3.67GYY156 pKa = 11.65YY157 pKa = 9.94PLAIEE162 pKa = 4.13IATLVEE168 pKa = 3.87QSRR171 pKa = 11.84TIINHH176 pKa = 5.58GLRR179 pKa = 11.84ATILKK184 pKa = 10.11RR185 pKa = 11.84CDD187 pKa = 4.45DD188 pKa = 3.75ISDD191 pKa = 4.37LEE193 pKa = 4.44LPSVDD198 pKa = 3.77LDD200 pKa = 3.71EE201 pKa = 6.11SIEE204 pKa = 4.48LISNSNIVSKK214 pKa = 10.9RR215 pKa = 11.84KK216 pKa = 5.88THH218 pKa = 6.42KK219 pKa = 10.1KK220 pKa = 8.83GKK222 pKa = 9.08KK223 pKa = 8.95RR224 pKa = 11.84KK225 pKa = 6.65NTKK228 pKa = 9.05TEE230 pKa = 4.07SEE232 pKa = 4.15SSEE235 pKa = 4.04FARR238 pKa = 11.84VPNFRR243 pKa = 11.84GNDD246 pKa = 3.41LSEE249 pKa = 4.94SGDD252 pKa = 3.6NEE254 pKa = 5.01DD255 pKa = 3.8II256 pKa = 4.75
MM1 pKa = 7.48ALVEE5 pKa = 4.33LKK7 pKa = 10.38EE8 pKa = 4.08PKK10 pKa = 9.52QLKK13 pKa = 10.49VNDD16 pKa = 3.94FVKK19 pKa = 10.43MSFADD24 pKa = 3.98KK25 pKa = 10.36ILPRR29 pKa = 11.84SLTRR33 pKa = 11.84LRR35 pKa = 11.84TVSISEE41 pKa = 4.26TNVVKK46 pKa = 10.9LSGLGSTVNLNILKK60 pKa = 10.4GVVLNSEE67 pKa = 4.39SKK69 pKa = 9.31YY70 pKa = 7.4VTIRR74 pKa = 11.84GVVISGVWMVPEE86 pKa = 4.63GCGGGATVTLMDD98 pKa = 4.15RR99 pKa = 11.84RR100 pKa = 11.84MKK102 pKa = 10.6GFKK105 pKa = 9.92NGLVAEE111 pKa = 5.18FKK113 pKa = 10.21TRR115 pKa = 11.84ASSRR119 pKa = 11.84DD120 pKa = 3.52FQFKK124 pKa = 10.15FIPNYY129 pKa = 10.04SMCVDD134 pKa = 3.75DD135 pKa = 5.38VKK137 pKa = 10.74RR138 pKa = 11.84APWEE142 pKa = 3.88LFFKK146 pKa = 10.93LVGVPIEE153 pKa = 4.42DD154 pKa = 3.67GYY156 pKa = 11.65YY157 pKa = 9.94PLAIEE162 pKa = 4.13IATLVEE168 pKa = 3.87QSRR171 pKa = 11.84TIINHH176 pKa = 5.58GLRR179 pKa = 11.84ATILKK184 pKa = 10.11RR185 pKa = 11.84CDD187 pKa = 4.45DD188 pKa = 3.75ISDD191 pKa = 4.37LEE193 pKa = 4.44LPSVDD198 pKa = 3.77LDD200 pKa = 3.71EE201 pKa = 6.11SIEE204 pKa = 4.48LISNSNIVSKK214 pKa = 10.9RR215 pKa = 11.84KK216 pKa = 5.88THH218 pKa = 6.42KK219 pKa = 10.1KK220 pKa = 8.83GKK222 pKa = 9.08KK223 pKa = 8.95RR224 pKa = 11.84KK225 pKa = 6.65NTKK228 pKa = 9.05TEE230 pKa = 4.07SEE232 pKa = 4.15SSEE235 pKa = 4.04FARR238 pKa = 11.84VPNFRR243 pKa = 11.84GNDD246 pKa = 3.41LSEE249 pKa = 4.94SGDD252 pKa = 3.6NEE254 pKa = 5.01DD255 pKa = 3.8II256 pKa = 4.75
Molecular weight: 28.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3226 |
174 |
1650 |
806.5 |
91.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.859 ± 0.329 | 2.449 ± 0.388 |
5.952 ± 0.154 | 6.355 ± 0.125 |
4.867 ± 0.157 | 4.898 ± 0.417 |
2.418 ± 0.393 | 5.394 ± 0.301 |
7.347 ± 0.456 | 9.423 ± 0.135 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.418 ± 0.182 | 4.278 ± 0.222 |
3.689 ± 0.134 | 2.635 ± 0.207 |
4.681 ± 0.404 | 8.772 ± 0.167 |
6.014 ± 0.467 | 7.812 ± 0.295 |
1.085 ± 0.053 | 3.627 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
