
Geomicrobium sp. JCM 19038
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillales incertae sedis; Geomicrobium; unclassified Geomicrobium
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
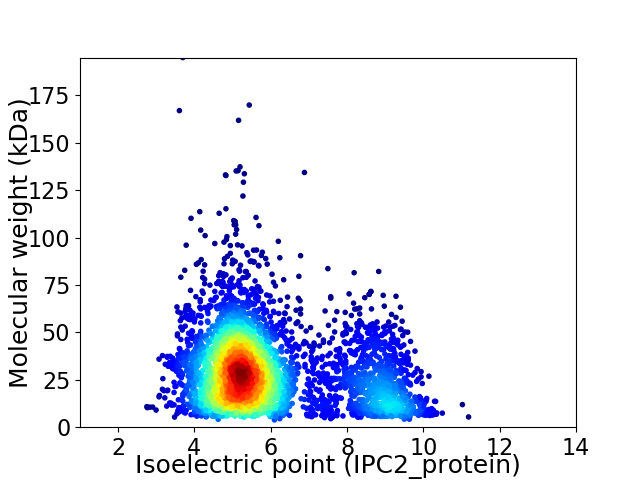
Virtual 2D-PAGE plot for 4086 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A061P1C4|A0A061P1C4_9BACL D-arabino-3-hexulose 6-phosphate formaldehyde lyase OS=Geomicrobium sp. JCM 19038 OX=1460635 GN=JCM19038_3996 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84GRR4 pKa = 11.84IVRR7 pKa = 11.84YY8 pKa = 10.06SKK10 pKa = 10.14ATALAGLALTATLTISSEE28 pKa = 3.67VDD30 pKa = 2.97AEE32 pKa = 4.02QGEE35 pKa = 4.7VLKK38 pKa = 10.65ILHH41 pKa = 6.49TNDD44 pKa = 2.15IHH46 pKa = 6.95ATFDD50 pKa = 3.61NFGKK54 pKa = 10.45AAGYY58 pKa = 9.49IDD60 pKa = 4.37QVRR63 pKa = 11.84EE64 pKa = 4.02DD65 pKa = 4.31YY66 pKa = 11.67DD67 pKa = 3.51NVLYY71 pKa = 10.96LDD73 pKa = 4.17AGDD76 pKa = 3.93YY77 pKa = 11.25ASGNPVVDD85 pKa = 4.73LNYY88 pKa = 10.0GIPMVEE94 pKa = 4.62AFNEE98 pKa = 4.02ANLDD102 pKa = 3.48VLTIGNHH109 pKa = 4.81EE110 pKa = 4.21FDD112 pKa = 3.97YY113 pKa = 11.34GQDD116 pKa = 3.31HH117 pKa = 7.77LINNMDD123 pKa = 3.58TSTASWLGANVDD135 pKa = 3.81TGSTGVDD142 pKa = 3.04NPEE145 pKa = 4.55PYY147 pKa = 10.92TMIDD151 pKa = 3.35LDD153 pKa = 4.2GVSVAILGITQAPPATAPANVEE175 pKa = 3.85GMTFDD180 pKa = 3.4SDD182 pKa = 4.12YY183 pKa = 11.2AAVAAAYY190 pKa = 9.62EE191 pKa = 4.21EE192 pKa = 4.67EE193 pKa = 4.58LTSEE197 pKa = 4.24ADD199 pKa = 3.51VIIALTHH206 pKa = 5.63IGYY209 pKa = 9.8NEE211 pKa = 3.86DD212 pKa = 3.32RR213 pKa = 11.84QLAEE217 pKa = 4.19DD218 pKa = 3.36VDD220 pKa = 4.35YY221 pKa = 11.18FDD223 pKa = 5.9VIIGGHH229 pKa = 5.08SHH231 pKa = 5.68TTLNSPQIVNGTLIVQTGSNLNNLGEE257 pKa = 4.13LTLAIEE263 pKa = 4.32NNEE266 pKa = 3.86VTEE269 pKa = 4.16LDD271 pKa = 3.62GTLVPVADD279 pKa = 5.02LDD281 pKa = 4.06TMNEE285 pKa = 3.96DD286 pKa = 3.18VQAVIDD292 pKa = 4.34HH293 pKa = 6.51YY294 pKa = 10.9NSEE297 pKa = 4.02MDD299 pKa = 3.42EE300 pKa = 4.21VLGEE304 pKa = 4.19VIGYY308 pKa = 8.59SEE310 pKa = 4.79NGLSRR315 pKa = 11.84DD316 pKa = 3.67GRR318 pKa = 11.84FQQDD322 pKa = 2.91APLGNFWTDD331 pKa = 3.9AMRR334 pKa = 11.84EE335 pKa = 3.85WAEE338 pKa = 3.63ADD340 pKa = 3.59VAFTNNGGIRR350 pKa = 11.84DD351 pKa = 4.66SIPSGDD357 pKa = 3.38VTLNEE362 pKa = 4.69IYY364 pKa = 10.31TIEE367 pKa = 4.22PFANEE372 pKa = 3.24IMTIEE377 pKa = 4.07MTGEE381 pKa = 4.17AIGNVLEE388 pKa = 4.29YY389 pKa = 10.55SYY391 pKa = 11.73SRR393 pKa = 11.84DD394 pKa = 2.8GRR396 pKa = 11.84NQIDD400 pKa = 3.92LQVSGMEE407 pKa = 4.05YY408 pKa = 10.48EE409 pKa = 4.58ILTGMTGQLSDD420 pKa = 3.78VNMTIDD426 pKa = 4.29GSPLEE431 pKa = 4.99DD432 pKa = 3.35EE433 pKa = 4.33EE434 pKa = 5.39TYY436 pKa = 10.23TVAVADD442 pKa = 4.2YY443 pKa = 9.69IGTGGSGYY451 pKa = 9.55EE452 pKa = 4.1FEE454 pKa = 5.5GEE456 pKa = 4.37VIQEE460 pKa = 4.3TVGFMTDD467 pKa = 5.5AMTDD471 pKa = 3.14WARR474 pKa = 11.84TMTDD478 pKa = 2.5QGEE481 pKa = 4.38TLDD484 pKa = 3.86YY485 pKa = 11.28ASGEE489 pKa = 4.29RR490 pKa = 11.84ISITIDD496 pKa = 3.08PSGPIPGTIIGSTEE510 pKa = 3.67TGLYY514 pKa = 9.0SQNKK518 pKa = 8.27AFQDD522 pKa = 3.51VGLGNLYY529 pKa = 9.71TDD531 pKa = 5.4SILDD535 pKa = 3.64DD536 pKa = 3.7TDD538 pKa = 4.59ADD540 pKa = 4.13FAFLNASSVTGQIPPGEE557 pKa = 4.12ITDD560 pKa = 3.93EE561 pKa = 4.1QIEE564 pKa = 4.14ALDD567 pKa = 3.8GFGNEE572 pKa = 3.99VVVVEE577 pKa = 4.44TTGEE581 pKa = 4.07RR582 pKa = 11.84IEE584 pKa = 4.25EE585 pKa = 4.35VILEE589 pKa = 4.01QSNYY593 pKa = 8.84HH594 pKa = 5.89QGVDD598 pKa = 3.8LQVSGLSYY606 pKa = 10.37TLIPEE611 pKa = 4.1EE612 pKa = 4.4SGSGFEE618 pKa = 4.66SVDD621 pKa = 2.81IVLPDD626 pKa = 3.64GSEE629 pKa = 4.09LEE631 pKa = 3.89EE632 pKa = 5.62DD633 pKa = 3.42EE634 pKa = 5.37AYY636 pKa = 10.66VVAYY640 pKa = 10.59NDD642 pKa = 3.69FMHH645 pKa = 6.95SNSFYY650 pKa = 11.23NFGEE654 pKa = 4.35LIEE657 pKa = 4.46EE658 pKa = 4.9KK659 pKa = 10.89GPVWQKK665 pKa = 9.9VVDD668 pKa = 4.1YY669 pKa = 9.73VTEE672 pKa = 3.87QNRR675 pKa = 11.84PINYY679 pKa = 9.62EE680 pKa = 3.45EE681 pKa = 4.4GLRR684 pKa = 11.84ITIEE688 pKa = 4.29GQDD691 pKa = 3.86VEE693 pKa = 4.6PDD695 pKa = 3.44PTGTWTVAEE704 pKa = 4.23ALQNNHH710 pKa = 5.57GEE712 pKa = 4.23RR713 pKa = 11.84AVRR716 pKa = 11.84GYY718 pKa = 10.43IIGSIQNNEE727 pKa = 3.77VVLGIGEE734 pKa = 4.32HH735 pKa = 6.33APSNLLLADD744 pKa = 4.96DD745 pKa = 4.88PNEE748 pKa = 3.98TDD750 pKa = 3.49RR751 pKa = 11.84SNMLPVQLVNQTPIRR766 pKa = 11.84NGLNLVDD773 pKa = 5.58HH774 pKa = 7.54PDD776 pKa = 3.47HH777 pKa = 6.21LQKK780 pKa = 10.82EE781 pKa = 4.71VVITGSLEE789 pKa = 4.21RR790 pKa = 11.84YY791 pKa = 9.22FGTPGMRR798 pKa = 11.84DD799 pKa = 3.26ANHH802 pKa = 6.3FAWVEE807 pKa = 3.81EE808 pKa = 4.34DD809 pKa = 4.81AVSCQYY815 pKa = 9.65DD816 pKa = 3.06TWQQSDD822 pKa = 4.71VYY824 pKa = 10.11TKK826 pKa = 10.88GDD828 pKa = 3.35RR829 pKa = 11.84VTHH832 pKa = 6.34NDD834 pKa = 2.81VVYY837 pKa = 9.71EE838 pKa = 4.16AKK840 pKa = 9.24WWTVGEE846 pKa = 4.06NPEE849 pKa = 4.62NSSEE853 pKa = 3.81WDD855 pKa = 3.17VWQVVADD862 pKa = 4.11CQSDD866 pKa = 3.48EE867 pKa = 4.54GDD869 pKa = 3.76GIKK872 pKa = 10.42NGRR875 pKa = 11.84RR876 pKa = 11.84RR877 pKa = 3.42
MM1 pKa = 7.9RR2 pKa = 11.84GRR4 pKa = 11.84IVRR7 pKa = 11.84YY8 pKa = 10.06SKK10 pKa = 10.14ATALAGLALTATLTISSEE28 pKa = 3.67VDD30 pKa = 2.97AEE32 pKa = 4.02QGEE35 pKa = 4.7VLKK38 pKa = 10.65ILHH41 pKa = 6.49TNDD44 pKa = 2.15IHH46 pKa = 6.95ATFDD50 pKa = 3.61NFGKK54 pKa = 10.45AAGYY58 pKa = 9.49IDD60 pKa = 4.37QVRR63 pKa = 11.84EE64 pKa = 4.02DD65 pKa = 4.31YY66 pKa = 11.67DD67 pKa = 3.51NVLYY71 pKa = 10.96LDD73 pKa = 4.17AGDD76 pKa = 3.93YY77 pKa = 11.25ASGNPVVDD85 pKa = 4.73LNYY88 pKa = 10.0GIPMVEE94 pKa = 4.62AFNEE98 pKa = 4.02ANLDD102 pKa = 3.48VLTIGNHH109 pKa = 4.81EE110 pKa = 4.21FDD112 pKa = 3.97YY113 pKa = 11.34GQDD116 pKa = 3.31HH117 pKa = 7.77LINNMDD123 pKa = 3.58TSTASWLGANVDD135 pKa = 3.81TGSTGVDD142 pKa = 3.04NPEE145 pKa = 4.55PYY147 pKa = 10.92TMIDD151 pKa = 3.35LDD153 pKa = 4.2GVSVAILGITQAPPATAPANVEE175 pKa = 3.85GMTFDD180 pKa = 3.4SDD182 pKa = 4.12YY183 pKa = 11.2AAVAAAYY190 pKa = 9.62EE191 pKa = 4.21EE192 pKa = 4.67EE193 pKa = 4.58LTSEE197 pKa = 4.24ADD199 pKa = 3.51VIIALTHH206 pKa = 5.63IGYY209 pKa = 9.8NEE211 pKa = 3.86DD212 pKa = 3.32RR213 pKa = 11.84QLAEE217 pKa = 4.19DD218 pKa = 3.36VDD220 pKa = 4.35YY221 pKa = 11.18FDD223 pKa = 5.9VIIGGHH229 pKa = 5.08SHH231 pKa = 5.68TTLNSPQIVNGTLIVQTGSNLNNLGEE257 pKa = 4.13LTLAIEE263 pKa = 4.32NNEE266 pKa = 3.86VTEE269 pKa = 4.16LDD271 pKa = 3.62GTLVPVADD279 pKa = 5.02LDD281 pKa = 4.06TMNEE285 pKa = 3.96DD286 pKa = 3.18VQAVIDD292 pKa = 4.34HH293 pKa = 6.51YY294 pKa = 10.9NSEE297 pKa = 4.02MDD299 pKa = 3.42EE300 pKa = 4.21VLGEE304 pKa = 4.19VIGYY308 pKa = 8.59SEE310 pKa = 4.79NGLSRR315 pKa = 11.84DD316 pKa = 3.67GRR318 pKa = 11.84FQQDD322 pKa = 2.91APLGNFWTDD331 pKa = 3.9AMRR334 pKa = 11.84EE335 pKa = 3.85WAEE338 pKa = 3.63ADD340 pKa = 3.59VAFTNNGGIRR350 pKa = 11.84DD351 pKa = 4.66SIPSGDD357 pKa = 3.38VTLNEE362 pKa = 4.69IYY364 pKa = 10.31TIEE367 pKa = 4.22PFANEE372 pKa = 3.24IMTIEE377 pKa = 4.07MTGEE381 pKa = 4.17AIGNVLEE388 pKa = 4.29YY389 pKa = 10.55SYY391 pKa = 11.73SRR393 pKa = 11.84DD394 pKa = 2.8GRR396 pKa = 11.84NQIDD400 pKa = 3.92LQVSGMEE407 pKa = 4.05YY408 pKa = 10.48EE409 pKa = 4.58ILTGMTGQLSDD420 pKa = 3.78VNMTIDD426 pKa = 4.29GSPLEE431 pKa = 4.99DD432 pKa = 3.35EE433 pKa = 4.33EE434 pKa = 5.39TYY436 pKa = 10.23TVAVADD442 pKa = 4.2YY443 pKa = 9.69IGTGGSGYY451 pKa = 9.55EE452 pKa = 4.1FEE454 pKa = 5.5GEE456 pKa = 4.37VIQEE460 pKa = 4.3TVGFMTDD467 pKa = 5.5AMTDD471 pKa = 3.14WARR474 pKa = 11.84TMTDD478 pKa = 2.5QGEE481 pKa = 4.38TLDD484 pKa = 3.86YY485 pKa = 11.28ASGEE489 pKa = 4.29RR490 pKa = 11.84ISITIDD496 pKa = 3.08PSGPIPGTIIGSTEE510 pKa = 3.67TGLYY514 pKa = 9.0SQNKK518 pKa = 8.27AFQDD522 pKa = 3.51VGLGNLYY529 pKa = 9.71TDD531 pKa = 5.4SILDD535 pKa = 3.64DD536 pKa = 3.7TDD538 pKa = 4.59ADD540 pKa = 4.13FAFLNASSVTGQIPPGEE557 pKa = 4.12ITDD560 pKa = 3.93EE561 pKa = 4.1QIEE564 pKa = 4.14ALDD567 pKa = 3.8GFGNEE572 pKa = 3.99VVVVEE577 pKa = 4.44TTGEE581 pKa = 4.07RR582 pKa = 11.84IEE584 pKa = 4.25EE585 pKa = 4.35VILEE589 pKa = 4.01QSNYY593 pKa = 8.84HH594 pKa = 5.89QGVDD598 pKa = 3.8LQVSGLSYY606 pKa = 10.37TLIPEE611 pKa = 4.1EE612 pKa = 4.4SGSGFEE618 pKa = 4.66SVDD621 pKa = 2.81IVLPDD626 pKa = 3.64GSEE629 pKa = 4.09LEE631 pKa = 3.89EE632 pKa = 5.62DD633 pKa = 3.42EE634 pKa = 5.37AYY636 pKa = 10.66VVAYY640 pKa = 10.59NDD642 pKa = 3.69FMHH645 pKa = 6.95SNSFYY650 pKa = 11.23NFGEE654 pKa = 4.35LIEE657 pKa = 4.46EE658 pKa = 4.9KK659 pKa = 10.89GPVWQKK665 pKa = 9.9VVDD668 pKa = 4.1YY669 pKa = 9.73VTEE672 pKa = 3.87QNRR675 pKa = 11.84PINYY679 pKa = 9.62EE680 pKa = 3.45EE681 pKa = 4.4GLRR684 pKa = 11.84ITIEE688 pKa = 4.29GQDD691 pKa = 3.86VEE693 pKa = 4.6PDD695 pKa = 3.44PTGTWTVAEE704 pKa = 4.23ALQNNHH710 pKa = 5.57GEE712 pKa = 4.23RR713 pKa = 11.84AVRR716 pKa = 11.84GYY718 pKa = 10.43IIGSIQNNEE727 pKa = 3.77VVLGIGEE734 pKa = 4.32HH735 pKa = 6.33APSNLLLADD744 pKa = 4.96DD745 pKa = 4.88PNEE748 pKa = 3.98TDD750 pKa = 3.49RR751 pKa = 11.84SNMLPVQLVNQTPIRR766 pKa = 11.84NGLNLVDD773 pKa = 5.58HH774 pKa = 7.54PDD776 pKa = 3.47HH777 pKa = 6.21LQKK780 pKa = 10.82EE781 pKa = 4.71VVITGSLEE789 pKa = 4.21RR790 pKa = 11.84YY791 pKa = 9.22FGTPGMRR798 pKa = 11.84DD799 pKa = 3.26ANHH802 pKa = 6.3FAWVEE807 pKa = 3.81EE808 pKa = 4.34DD809 pKa = 4.81AVSCQYY815 pKa = 9.65DD816 pKa = 3.06TWQQSDD822 pKa = 4.71VYY824 pKa = 10.11TKK826 pKa = 10.88GDD828 pKa = 3.35RR829 pKa = 11.84VTHH832 pKa = 6.34NDD834 pKa = 2.81VVYY837 pKa = 9.71EE838 pKa = 4.16AKK840 pKa = 9.24WWTVGEE846 pKa = 4.06NPEE849 pKa = 4.62NSSEE853 pKa = 3.81WDD855 pKa = 3.17VWQVVADD862 pKa = 4.11CQSDD866 pKa = 3.48EE867 pKa = 4.54GDD869 pKa = 3.76GIKK872 pKa = 10.42NGRR875 pKa = 11.84RR876 pKa = 11.84RR877 pKa = 3.42
Molecular weight: 95.99 kDa
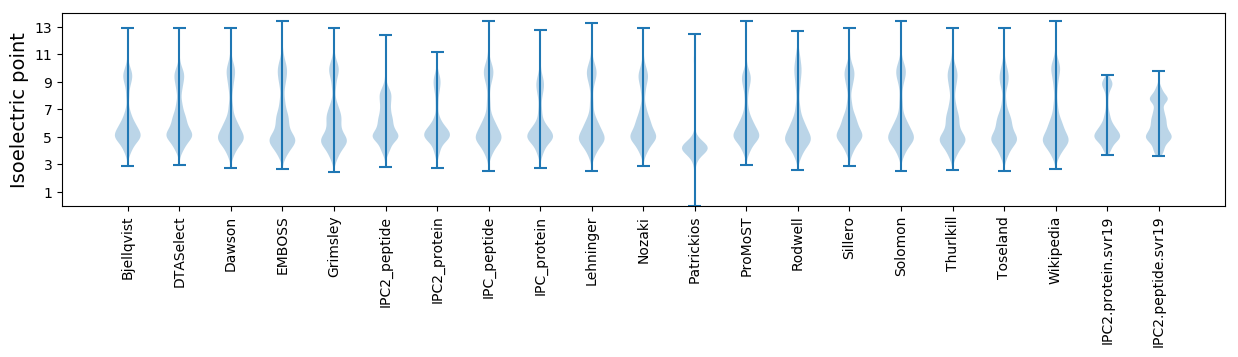
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A061NWW3|A0A061NWW3_9BACL Transcriptional regulator DeoR family OS=Geomicrobium sp. JCM 19038 OX=1460635 GN=JCM19038_2974 PE=4 SV=1
MM1 pKa = 7.38SKK3 pKa = 8.63KK4 pKa = 8.77TFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.02KK15 pKa = 8.47VHH17 pKa = 5.57GFRR20 pKa = 11.84SRR22 pKa = 11.84MSTKK26 pKa = 10.33NGRR29 pKa = 11.84LVLKK33 pKa = 10.39RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 10.33GRR40 pKa = 11.84KK41 pKa = 8.52VLSAA45 pKa = 4.05
MM1 pKa = 7.38SKK3 pKa = 8.63KK4 pKa = 8.77TFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.02KK15 pKa = 8.47VHH17 pKa = 5.57GFRR20 pKa = 11.84SRR22 pKa = 11.84MSTKK26 pKa = 10.33NGRR29 pKa = 11.84LVLKK33 pKa = 10.39RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 10.33GRR40 pKa = 11.84KK41 pKa = 8.52VLSAA45 pKa = 4.05
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1114903 |
37 |
1794 |
272.9 |
30.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.444 ± 0.037 | 0.625 ± 0.012 |
5.485 ± 0.037 | 7.777 ± 0.051 |
4.346 ± 0.032 | 6.874 ± 0.034 |
2.461 ± 0.022 | 7.082 ± 0.035 |
5.165 ± 0.04 | 9.683 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.914 ± 0.019 | 3.946 ± 0.026 |
3.661 ± 0.027 | 4.044 ± 0.025 |
4.539 ± 0.028 | 6.113 ± 0.026 |
5.915 ± 0.028 | 7.514 ± 0.032 |
1.005 ± 0.015 | 3.407 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
