
Bacteroidetes bacterium SCGC AAA795-G10
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; unclassified Bacteroidetes
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
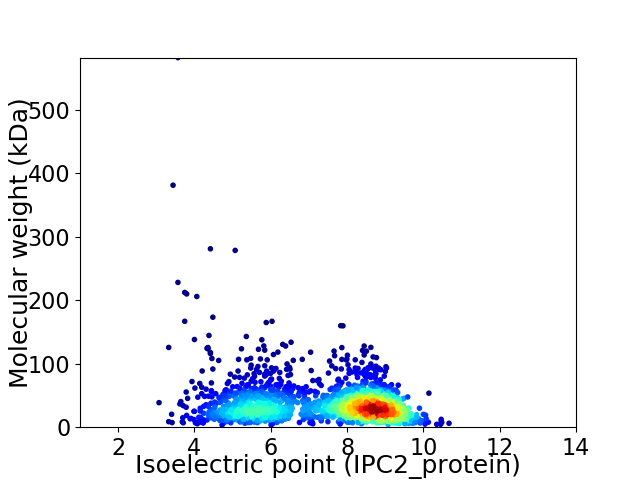
Virtual 2D-PAGE plot for 1767 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
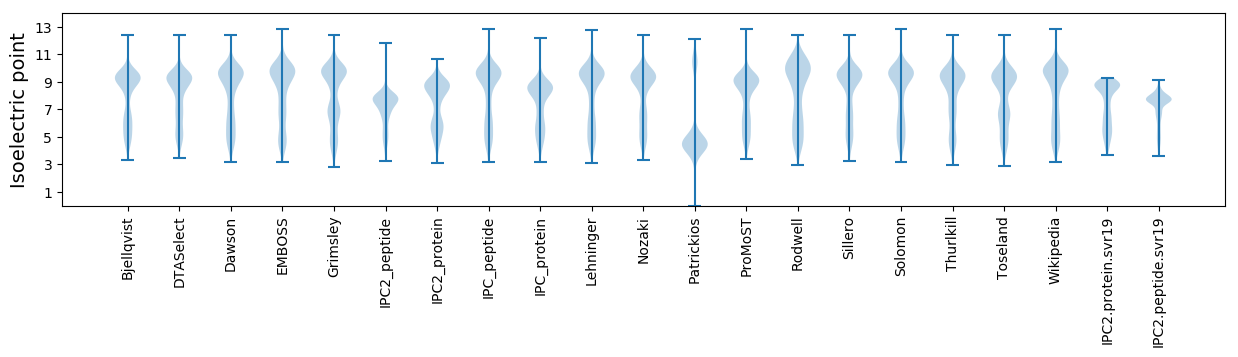
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U2S0H3|A0A2U2S0H3_9BACT Uncharacterized protein OS=Bacteroidetes bacterium SCGC AAA795-G10 OX=2175803 GN=DEJ39_01680 PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 9.83EE3 pKa = 3.98VKK5 pKa = 10.35SLLFCLLSIPSFIFGQIVTDD25 pKa = 4.19GLILNLDD32 pKa = 3.76TRR34 pKa = 11.84DD35 pKa = 3.62SNSYY39 pKa = 10.17SGSGNTWNDD48 pKa = 2.6ISGNNYY54 pKa = 9.28NATIVGGTSFSNNQFTLDD72 pKa = 3.44GSNNYY77 pKa = 9.34ISTPAIPSVGISTQDD92 pKa = 3.11YY93 pKa = 8.36SLEE96 pKa = 4.31VIVTPTSTSGNVINMSNQSDD116 pKa = 3.69NLGWNMPPLASANSTFYY133 pKa = 11.27AQLHH137 pKa = 5.78SSANGVTNGISSSFNVNQKK156 pKa = 9.58YY157 pKa = 10.39HH158 pKa = 6.91LVLTWDD164 pKa = 3.51YY165 pKa = 11.59DD166 pKa = 3.75ANANNRR172 pKa = 11.84VFKK175 pKa = 10.84LYY177 pKa = 10.91VNNVLVGQRR186 pKa = 11.84SNITYY191 pKa = 10.44SSSGSNNYY199 pKa = 9.1FFLGKK204 pKa = 9.98DD205 pKa = 3.33NPGCCDD211 pKa = 3.05EE212 pKa = 4.49FNGVEE217 pKa = 4.09SRR219 pKa = 11.84DD220 pKa = 3.56FAGKK224 pKa = 9.55YY225 pKa = 7.41EE226 pKa = 3.98AFRR229 pKa = 11.84VYY231 pKa = 10.82SKK233 pKa = 11.13ALSAQEE239 pKa = 3.8VAQNYY244 pKa = 7.38TSSSNQTTTTLSISNEE260 pKa = 3.39TRR262 pKa = 11.84TFDD265 pKa = 3.6EE266 pKa = 5.08LNFNLSPTSNSSQAFNYY283 pKa = 10.04LISDD287 pKa = 3.96TNVATVNGNTVTIVGVGTTSVTVSQAAGGGYY318 pKa = 7.43TAATATMTLTINKK331 pKa = 7.15GTPTISFSNLTKK343 pKa = 10.38TYY345 pKa = 10.88GDD347 pKa = 3.8ANFDD351 pKa = 3.71LSATSSSTGGLTYY364 pKa = 10.58SIADD368 pKa = 3.58ANIASLSVSNVTIVGAGTTSITVTQAADD396 pKa = 3.16ANYY399 pKa = 10.68NSATSTITLTVSKK412 pKa = 10.67LNTTITLSNITKK424 pKa = 10.26SALDD428 pKa = 3.42ADD430 pKa = 4.6FNLDD434 pKa = 4.21AISSRR439 pKa = 11.84PGTQVVTSGLVLHH452 pKa = 7.01LDD454 pKa = 3.69ASDD457 pKa = 3.37TDD459 pKa = 4.46SYY461 pKa = 12.04DD462 pKa = 3.43SASGGNTWYY471 pKa = 10.37DD472 pKa = 2.85ISGNSNDD479 pKa = 3.03FTLYY483 pKa = 10.82GNPTFRR489 pKa = 11.84SDD491 pKa = 3.55LNSGTFEE498 pKa = 3.82FDD500 pKa = 3.18EE501 pKa = 4.76NNDD504 pKa = 3.62YY505 pKa = 11.15AQSNSTTVLNSTQYY519 pKa = 10.44TKK521 pKa = 10.6IAVYY525 pKa = 10.01YY526 pKa = 9.07PKK528 pKa = 10.67SKK530 pKa = 9.68TKK532 pKa = 10.81NIVSGGNEE540 pKa = 3.65AAHH543 pKa = 6.39AFWMANSTNTIRR555 pKa = 11.84AGHH558 pKa = 5.22NTSWSQVSYY567 pKa = 11.38SPGNMLNSWHH577 pKa = 6.18YY578 pKa = 10.62SAVSFSNSDD587 pKa = 3.39GFKK590 pKa = 10.62LYY592 pKa = 11.25YY593 pKa = 10.21NGNLVDD599 pKa = 5.06SDD601 pKa = 4.1NFTNTPNGNSPGLVRR616 pKa = 11.84IASYY620 pKa = 11.24NGGNLFDD627 pKa = 4.91GYY629 pKa = 10.63IPVVLIYY636 pKa = 10.95NRR638 pKa = 11.84VLTSSEE644 pKa = 3.41IQNNYY649 pKa = 9.38NNLAQRR655 pKa = 11.84YY656 pKa = 8.32GLMNIGNYY664 pKa = 10.52SNVDD668 pKa = 3.62LTYY671 pKa = 10.72SIADD675 pKa = 3.56TSIATLNGNKK685 pKa = 9.59VSIVGAGTTTITVSQAADD703 pKa = 3.42SNFNSGSKK711 pKa = 9.67TITLTVNTVTPTITFSNEE729 pKa = 2.81SRR731 pKa = 11.84TFGDD735 pKa = 3.55SNFDD739 pKa = 3.62FSTTSNSSGTFSYY752 pKa = 10.39IISDD756 pKa = 3.77TNIATVSGNNVTIRR770 pKa = 11.84GAGTTSVTVIQAANGIYY787 pKa = 10.02SSGVATMTLTINKK800 pKa = 9.26ADD802 pKa = 3.73PTFSFSDD809 pKa = 3.47LTKK812 pKa = 10.69TFGDD816 pKa = 3.87PNFDD820 pKa = 4.17LSTSSNSNGIITFTISNTSVARR842 pKa = 11.84LNPITGGLLLHH853 pKa = 6.58VDD855 pKa = 3.41SRR857 pKa = 11.84DD858 pKa = 3.43PTSYY862 pKa = 10.02PGSGNTVYY870 pKa = 10.88DD871 pKa = 3.9LSGNNHH877 pKa = 7.15DD878 pKa = 4.38FTISGNMNYY887 pKa = 10.7DD888 pKa = 3.23NTNGFTFEE896 pKa = 4.19RR897 pKa = 11.84GQTAKK902 pKa = 10.77YY903 pKa = 9.93LKK905 pKa = 8.73QTNFPHH911 pKa = 6.48PTTTITNEE919 pKa = 3.85ILVKK923 pKa = 10.22TSSNDD928 pKa = 3.25NAGLLSYY935 pKa = 11.32NKK937 pKa = 9.32TDD939 pKa = 3.46NDD941 pKa = 3.75NANLIYY947 pKa = 9.49ITSGQVRR954 pKa = 11.84THH956 pKa = 5.98HH957 pKa = 6.06TPGFNRR963 pKa = 11.84NTNVNLSDD971 pKa = 4.02NQWHH975 pKa = 6.67HH976 pKa = 6.61FVQTIDD982 pKa = 3.31KK983 pKa = 10.65SSGVDD988 pKa = 3.4KK989 pKa = 10.77IYY991 pKa = 11.01LDD993 pKa = 3.82GVLVLTYY1000 pKa = 8.92NTNNTLIPTNGCLAIGQDD1018 pKa = 4.04FDD1020 pKa = 4.11TTCGGFNANDD1030 pKa = 3.5AFGGYY1035 pKa = 9.91LPLVRR1040 pKa = 11.84MYY1042 pKa = 11.37DD1043 pKa = 3.66RR1044 pKa = 11.84ILTAEE1049 pKa = 4.22EE1050 pKa = 3.9VAQNYY1055 pKa = 9.66NSLSGATGTGSNTRR1069 pKa = 11.84IVGAGQTVITFTQAATNNYY1088 pKa = 8.8NAVTASMTLTVNKK1101 pKa = 8.12ATPTISISDD1110 pKa = 3.72EE1111 pKa = 3.5NRR1113 pKa = 11.84TFGDD1117 pKa = 3.36PAFNLSATSSSTGAFTYY1134 pKa = 9.72TIADD1138 pKa = 3.66ANVATVIGNTVTIEE1152 pKa = 4.23GGGTTSVTVTQAADD1166 pKa = 3.42SNYY1169 pKa = 10.55SSATSTITLTVNKK1182 pKa = 9.47FDD1184 pKa = 3.53PTIIFTGVVTRR1195 pKa = 11.84TYY1197 pKa = 11.26NDD1199 pKa = 3.03SDD1201 pKa = 4.19FNLSALSSSTASFTYY1216 pKa = 10.36SIADD1220 pKa = 3.33ASVATVSGNTVTIVGAGTTTITVSQLANANFNSATASASLIISKK1264 pKa = 10.44DD1265 pKa = 3.39NPTLTNFNPIVRR1277 pKa = 11.84TFGDD1281 pKa = 3.22ASFEE1285 pKa = 4.12IVEE1288 pKa = 4.2PTKK1291 pKa = 11.19NGDD1294 pKa = 3.31NTGSFSYY1301 pKa = 10.25TLLNQNIANISGNTVSITNSGQTSITATLSADD1333 pKa = 3.33NNYY1336 pKa = 10.49KK1337 pKa = 10.54SGTISTSFVVNKK1349 pKa = 9.85KK1350 pKa = 9.52NQTITVGALPTTKK1363 pKa = 9.8PLKK1366 pKa = 10.47DD1367 pKa = 3.57FTSIPITASSSSGSPIVISLSAGSAASVSGTVGNYY1402 pKa = 9.55EE1403 pKa = 4.38LVSIQQTGLVTITFTTDD1420 pKa = 2.87DD1421 pKa = 4.29SGNPNYY1427 pKa = 10.76NSATTTVVIDD1437 pKa = 3.96VVKK1440 pKa = 9.71TNQNISFNTTPTTQLTYY1457 pKa = 10.77TEE1459 pKa = 4.76NLDD1462 pKa = 3.62LTLDD1466 pKa = 3.82AVASSGLALSYY1477 pKa = 10.97SIVAGNNATLNNATLSFNDD1496 pKa = 3.48SGQLTVEE1503 pKa = 4.39ITQQGNNLYY1512 pKa = 9.41NQAVPVRR1519 pKa = 11.84VVFVVGQGNTSLSNFTIPTKK1539 pKa = 10.68TNLDD1543 pKa = 3.21NDD1545 pKa = 4.28FNLTPPTSNRR1555 pKa = 11.84SGNITYY1561 pKa = 10.47SSSNLNVATLSGTLISINGIGNTTITATQLANSRR1595 pKa = 11.84YY1596 pKa = 8.86KK1597 pKa = 10.45SASISAVFRR1606 pKa = 11.84VLVGDD1611 pKa = 3.76SDD1613 pKa = 4.53GDD1615 pKa = 4.04GLIDD1619 pKa = 4.71SQDD1622 pKa = 3.41NCPTVANPDD1631 pKa = 3.53QADD1634 pKa = 3.49NDD1636 pKa = 3.56QDD1638 pKa = 4.06GVGNVCDD1645 pKa = 3.53NCVDD1649 pKa = 3.83VNNPLQLDD1657 pKa = 4.23LDD1659 pKa = 4.15GDD1661 pKa = 4.69GYY1663 pKa = 11.58GDD1665 pKa = 3.58SCDD1668 pKa = 4.96AFTLDD1673 pKa = 4.0ASEE1676 pKa = 4.95HH1677 pKa = 6.39ADD1679 pKa = 3.53SDD1681 pKa = 3.99GDD1683 pKa = 4.58GIGDD1687 pKa = 4.38NADD1690 pKa = 3.46TDD1692 pKa = 4.28DD1693 pKa = 6.69DD1694 pKa = 4.33NDD1696 pKa = 3.47NWSDD1700 pKa = 3.82TIEE1703 pKa = 4.34IACGTNPRR1711 pKa = 11.84DD1712 pKa = 4.17SADD1715 pKa = 3.74KK1716 pKa = 10.86PIDD1719 pKa = 3.69SDD1721 pKa = 3.96NDD1723 pKa = 3.59GDD1725 pKa = 4.77PDD1727 pKa = 5.16CLDD1730 pKa = 4.67PDD1732 pKa = 4.94DD1733 pKa = 6.46DD1734 pKa = 4.54NDD1736 pKa = 4.53GYY1738 pKa = 11.65LDD1740 pKa = 3.94TEE1742 pKa = 4.63DD1743 pKa = 5.81LFPFDD1748 pKa = 3.68NQEE1751 pKa = 3.8WADD1754 pKa = 3.68NDD1756 pKa = 3.81LDD1758 pKa = 5.2GIGDD1762 pKa = 3.82NADD1765 pKa = 3.44TDD1767 pKa = 4.27DD1768 pKa = 6.91DD1769 pKa = 4.2NDD1771 pKa = 3.4QYY1773 pKa = 11.76LDD1775 pKa = 3.65QDD1777 pKa = 4.66EE1778 pKa = 4.98IDD1780 pKa = 4.53CLTDD1784 pKa = 3.78PFDD1787 pKa = 4.4SASTPDD1793 pKa = 5.51DD1794 pKa = 3.88FDD1796 pKa = 5.98KK1797 pKa = 11.44DD1798 pKa = 4.76LIPDD1802 pKa = 5.15CIDD1805 pKa = 3.96PNDD1808 pKa = 5.06DD1809 pKa = 4.3NDD1811 pKa = 4.17SCPDD1815 pKa = 3.6TEE1817 pKa = 6.26DD1818 pKa = 4.08EE1819 pKa = 5.09FPLDD1823 pKa = 4.56PEE1825 pKa = 4.75FCQDD1829 pKa = 3.17TDD1831 pKa = 3.7GDD1833 pKa = 4.53GIDD1836 pKa = 4.76DD1837 pKa = 4.5RR1838 pKa = 11.84FDD1840 pKa = 3.97FDD1842 pKa = 4.95SDD1844 pKa = 3.38NDD1846 pKa = 4.81GIPDD1850 pKa = 3.82HH1851 pKa = 7.37RR1852 pKa = 11.84DD1853 pKa = 3.17QFPQDD1858 pKa = 3.76PNASADD1864 pKa = 3.56GDD1866 pKa = 3.94GDD1868 pKa = 5.02GIPDD1872 pKa = 4.54SQDD1875 pKa = 2.67TDD1877 pKa = 3.55KK1878 pKa = 11.79NNDD1881 pKa = 3.52GFPDD1885 pKa = 3.63DD1886 pKa = 4.22QIIVSSVVTPNQPGIEE1902 pKa = 4.25EE1903 pKa = 4.03TWKK1906 pKa = 10.68VINIEE1911 pKa = 4.32DD1912 pKa = 3.69YY1913 pKa = 9.46PYY1915 pKa = 10.48TSVRR1919 pKa = 11.84VYY1921 pKa = 10.88APDD1924 pKa = 3.62GSRR1927 pKa = 11.84VFQSINYY1934 pKa = 8.31KK1935 pKa = 10.36NDD1937 pKa = 2.62WRR1939 pKa = 11.84GTNIRR1944 pKa = 11.84TGRR1947 pKa = 11.84PLPTGPYY1954 pKa = 8.88YY1955 pKa = 10.97YY1956 pKa = 10.11RR1957 pKa = 11.84IEE1959 pKa = 4.29LGGTSGEE1966 pKa = 4.78IIDD1969 pKa = 3.52GWLYY1973 pKa = 10.67IFNN1976 pKa = 4.53
MM1 pKa = 7.54KK2 pKa = 9.83EE3 pKa = 3.98VKK5 pKa = 10.35SLLFCLLSIPSFIFGQIVTDD25 pKa = 4.19GLILNLDD32 pKa = 3.76TRR34 pKa = 11.84DD35 pKa = 3.62SNSYY39 pKa = 10.17SGSGNTWNDD48 pKa = 2.6ISGNNYY54 pKa = 9.28NATIVGGTSFSNNQFTLDD72 pKa = 3.44GSNNYY77 pKa = 9.34ISTPAIPSVGISTQDD92 pKa = 3.11YY93 pKa = 8.36SLEE96 pKa = 4.31VIVTPTSTSGNVINMSNQSDD116 pKa = 3.69NLGWNMPPLASANSTFYY133 pKa = 11.27AQLHH137 pKa = 5.78SSANGVTNGISSSFNVNQKK156 pKa = 9.58YY157 pKa = 10.39HH158 pKa = 6.91LVLTWDD164 pKa = 3.51YY165 pKa = 11.59DD166 pKa = 3.75ANANNRR172 pKa = 11.84VFKK175 pKa = 10.84LYY177 pKa = 10.91VNNVLVGQRR186 pKa = 11.84SNITYY191 pKa = 10.44SSSGSNNYY199 pKa = 9.1FFLGKK204 pKa = 9.98DD205 pKa = 3.33NPGCCDD211 pKa = 3.05EE212 pKa = 4.49FNGVEE217 pKa = 4.09SRR219 pKa = 11.84DD220 pKa = 3.56FAGKK224 pKa = 9.55YY225 pKa = 7.41EE226 pKa = 3.98AFRR229 pKa = 11.84VYY231 pKa = 10.82SKK233 pKa = 11.13ALSAQEE239 pKa = 3.8VAQNYY244 pKa = 7.38TSSSNQTTTTLSISNEE260 pKa = 3.39TRR262 pKa = 11.84TFDD265 pKa = 3.6EE266 pKa = 5.08LNFNLSPTSNSSQAFNYY283 pKa = 10.04LISDD287 pKa = 3.96TNVATVNGNTVTIVGVGTTSVTVSQAAGGGYY318 pKa = 7.43TAATATMTLTINKK331 pKa = 7.15GTPTISFSNLTKK343 pKa = 10.38TYY345 pKa = 10.88GDD347 pKa = 3.8ANFDD351 pKa = 3.71LSATSSSTGGLTYY364 pKa = 10.58SIADD368 pKa = 3.58ANIASLSVSNVTIVGAGTTSITVTQAADD396 pKa = 3.16ANYY399 pKa = 10.68NSATSTITLTVSKK412 pKa = 10.67LNTTITLSNITKK424 pKa = 10.26SALDD428 pKa = 3.42ADD430 pKa = 4.6FNLDD434 pKa = 4.21AISSRR439 pKa = 11.84PGTQVVTSGLVLHH452 pKa = 7.01LDD454 pKa = 3.69ASDD457 pKa = 3.37TDD459 pKa = 4.46SYY461 pKa = 12.04DD462 pKa = 3.43SASGGNTWYY471 pKa = 10.37DD472 pKa = 2.85ISGNSNDD479 pKa = 3.03FTLYY483 pKa = 10.82GNPTFRR489 pKa = 11.84SDD491 pKa = 3.55LNSGTFEE498 pKa = 3.82FDD500 pKa = 3.18EE501 pKa = 4.76NNDD504 pKa = 3.62YY505 pKa = 11.15AQSNSTTVLNSTQYY519 pKa = 10.44TKK521 pKa = 10.6IAVYY525 pKa = 10.01YY526 pKa = 9.07PKK528 pKa = 10.67SKK530 pKa = 9.68TKK532 pKa = 10.81NIVSGGNEE540 pKa = 3.65AAHH543 pKa = 6.39AFWMANSTNTIRR555 pKa = 11.84AGHH558 pKa = 5.22NTSWSQVSYY567 pKa = 11.38SPGNMLNSWHH577 pKa = 6.18YY578 pKa = 10.62SAVSFSNSDD587 pKa = 3.39GFKK590 pKa = 10.62LYY592 pKa = 11.25YY593 pKa = 10.21NGNLVDD599 pKa = 5.06SDD601 pKa = 4.1NFTNTPNGNSPGLVRR616 pKa = 11.84IASYY620 pKa = 11.24NGGNLFDD627 pKa = 4.91GYY629 pKa = 10.63IPVVLIYY636 pKa = 10.95NRR638 pKa = 11.84VLTSSEE644 pKa = 3.41IQNNYY649 pKa = 9.38NNLAQRR655 pKa = 11.84YY656 pKa = 8.32GLMNIGNYY664 pKa = 10.52SNVDD668 pKa = 3.62LTYY671 pKa = 10.72SIADD675 pKa = 3.56TSIATLNGNKK685 pKa = 9.59VSIVGAGTTTITVSQAADD703 pKa = 3.42SNFNSGSKK711 pKa = 9.67TITLTVNTVTPTITFSNEE729 pKa = 2.81SRR731 pKa = 11.84TFGDD735 pKa = 3.55SNFDD739 pKa = 3.62FSTTSNSSGTFSYY752 pKa = 10.39IISDD756 pKa = 3.77TNIATVSGNNVTIRR770 pKa = 11.84GAGTTSVTVIQAANGIYY787 pKa = 10.02SSGVATMTLTINKK800 pKa = 9.26ADD802 pKa = 3.73PTFSFSDD809 pKa = 3.47LTKK812 pKa = 10.69TFGDD816 pKa = 3.87PNFDD820 pKa = 4.17LSTSSNSNGIITFTISNTSVARR842 pKa = 11.84LNPITGGLLLHH853 pKa = 6.58VDD855 pKa = 3.41SRR857 pKa = 11.84DD858 pKa = 3.43PTSYY862 pKa = 10.02PGSGNTVYY870 pKa = 10.88DD871 pKa = 3.9LSGNNHH877 pKa = 7.15DD878 pKa = 4.38FTISGNMNYY887 pKa = 10.7DD888 pKa = 3.23NTNGFTFEE896 pKa = 4.19RR897 pKa = 11.84GQTAKK902 pKa = 10.77YY903 pKa = 9.93LKK905 pKa = 8.73QTNFPHH911 pKa = 6.48PTTTITNEE919 pKa = 3.85ILVKK923 pKa = 10.22TSSNDD928 pKa = 3.25NAGLLSYY935 pKa = 11.32NKK937 pKa = 9.32TDD939 pKa = 3.46NDD941 pKa = 3.75NANLIYY947 pKa = 9.49ITSGQVRR954 pKa = 11.84THH956 pKa = 5.98HH957 pKa = 6.06TPGFNRR963 pKa = 11.84NTNVNLSDD971 pKa = 4.02NQWHH975 pKa = 6.67HH976 pKa = 6.61FVQTIDD982 pKa = 3.31KK983 pKa = 10.65SSGVDD988 pKa = 3.4KK989 pKa = 10.77IYY991 pKa = 11.01LDD993 pKa = 3.82GVLVLTYY1000 pKa = 8.92NTNNTLIPTNGCLAIGQDD1018 pKa = 4.04FDD1020 pKa = 4.11TTCGGFNANDD1030 pKa = 3.5AFGGYY1035 pKa = 9.91LPLVRR1040 pKa = 11.84MYY1042 pKa = 11.37DD1043 pKa = 3.66RR1044 pKa = 11.84ILTAEE1049 pKa = 4.22EE1050 pKa = 3.9VAQNYY1055 pKa = 9.66NSLSGATGTGSNTRR1069 pKa = 11.84IVGAGQTVITFTQAATNNYY1088 pKa = 8.8NAVTASMTLTVNKK1101 pKa = 8.12ATPTISISDD1110 pKa = 3.72EE1111 pKa = 3.5NRR1113 pKa = 11.84TFGDD1117 pKa = 3.36PAFNLSATSSSTGAFTYY1134 pKa = 9.72TIADD1138 pKa = 3.66ANVATVIGNTVTIEE1152 pKa = 4.23GGGTTSVTVTQAADD1166 pKa = 3.42SNYY1169 pKa = 10.55SSATSTITLTVNKK1182 pKa = 9.47FDD1184 pKa = 3.53PTIIFTGVVTRR1195 pKa = 11.84TYY1197 pKa = 11.26NDD1199 pKa = 3.03SDD1201 pKa = 4.19FNLSALSSSTASFTYY1216 pKa = 10.36SIADD1220 pKa = 3.33ASVATVSGNTVTIVGAGTTTITVSQLANANFNSATASASLIISKK1264 pKa = 10.44DD1265 pKa = 3.39NPTLTNFNPIVRR1277 pKa = 11.84TFGDD1281 pKa = 3.22ASFEE1285 pKa = 4.12IVEE1288 pKa = 4.2PTKK1291 pKa = 11.19NGDD1294 pKa = 3.31NTGSFSYY1301 pKa = 10.25TLLNQNIANISGNTVSITNSGQTSITATLSADD1333 pKa = 3.33NNYY1336 pKa = 10.49KK1337 pKa = 10.54SGTISTSFVVNKK1349 pKa = 9.85KK1350 pKa = 9.52NQTITVGALPTTKK1363 pKa = 9.8PLKK1366 pKa = 10.47DD1367 pKa = 3.57FTSIPITASSSSGSPIVISLSAGSAASVSGTVGNYY1402 pKa = 9.55EE1403 pKa = 4.38LVSIQQTGLVTITFTTDD1420 pKa = 2.87DD1421 pKa = 4.29SGNPNYY1427 pKa = 10.76NSATTTVVIDD1437 pKa = 3.96VVKK1440 pKa = 9.71TNQNISFNTTPTTQLTYY1457 pKa = 10.77TEE1459 pKa = 4.76NLDD1462 pKa = 3.62LTLDD1466 pKa = 3.82AVASSGLALSYY1477 pKa = 10.97SIVAGNNATLNNATLSFNDD1496 pKa = 3.48SGQLTVEE1503 pKa = 4.39ITQQGNNLYY1512 pKa = 9.41NQAVPVRR1519 pKa = 11.84VVFVVGQGNTSLSNFTIPTKK1539 pKa = 10.68TNLDD1543 pKa = 3.21NDD1545 pKa = 4.28FNLTPPTSNRR1555 pKa = 11.84SGNITYY1561 pKa = 10.47SSSNLNVATLSGTLISINGIGNTTITATQLANSRR1595 pKa = 11.84YY1596 pKa = 8.86KK1597 pKa = 10.45SASISAVFRR1606 pKa = 11.84VLVGDD1611 pKa = 3.76SDD1613 pKa = 4.53GDD1615 pKa = 4.04GLIDD1619 pKa = 4.71SQDD1622 pKa = 3.41NCPTVANPDD1631 pKa = 3.53QADD1634 pKa = 3.49NDD1636 pKa = 3.56QDD1638 pKa = 4.06GVGNVCDD1645 pKa = 3.53NCVDD1649 pKa = 3.83VNNPLQLDD1657 pKa = 4.23LDD1659 pKa = 4.15GDD1661 pKa = 4.69GYY1663 pKa = 11.58GDD1665 pKa = 3.58SCDD1668 pKa = 4.96AFTLDD1673 pKa = 4.0ASEE1676 pKa = 4.95HH1677 pKa = 6.39ADD1679 pKa = 3.53SDD1681 pKa = 3.99GDD1683 pKa = 4.58GIGDD1687 pKa = 4.38NADD1690 pKa = 3.46TDD1692 pKa = 4.28DD1693 pKa = 6.69DD1694 pKa = 4.33NDD1696 pKa = 3.47NWSDD1700 pKa = 3.82TIEE1703 pKa = 4.34IACGTNPRR1711 pKa = 11.84DD1712 pKa = 4.17SADD1715 pKa = 3.74KK1716 pKa = 10.86PIDD1719 pKa = 3.69SDD1721 pKa = 3.96NDD1723 pKa = 3.59GDD1725 pKa = 4.77PDD1727 pKa = 5.16CLDD1730 pKa = 4.67PDD1732 pKa = 4.94DD1733 pKa = 6.46DD1734 pKa = 4.54NDD1736 pKa = 4.53GYY1738 pKa = 11.65LDD1740 pKa = 3.94TEE1742 pKa = 4.63DD1743 pKa = 5.81LFPFDD1748 pKa = 3.68NQEE1751 pKa = 3.8WADD1754 pKa = 3.68NDD1756 pKa = 3.81LDD1758 pKa = 5.2GIGDD1762 pKa = 3.82NADD1765 pKa = 3.44TDD1767 pKa = 4.27DD1768 pKa = 6.91DD1769 pKa = 4.2NDD1771 pKa = 3.4QYY1773 pKa = 11.76LDD1775 pKa = 3.65QDD1777 pKa = 4.66EE1778 pKa = 4.98IDD1780 pKa = 4.53CLTDD1784 pKa = 3.78PFDD1787 pKa = 4.4SASTPDD1793 pKa = 5.51DD1794 pKa = 3.88FDD1796 pKa = 5.98KK1797 pKa = 11.44DD1798 pKa = 4.76LIPDD1802 pKa = 5.15CIDD1805 pKa = 3.96PNDD1808 pKa = 5.06DD1809 pKa = 4.3NDD1811 pKa = 4.17SCPDD1815 pKa = 3.6TEE1817 pKa = 6.26DD1818 pKa = 4.08EE1819 pKa = 5.09FPLDD1823 pKa = 4.56PEE1825 pKa = 4.75FCQDD1829 pKa = 3.17TDD1831 pKa = 3.7GDD1833 pKa = 4.53GIDD1836 pKa = 4.76DD1837 pKa = 4.5RR1838 pKa = 11.84FDD1840 pKa = 3.97FDD1842 pKa = 4.95SDD1844 pKa = 3.38NDD1846 pKa = 4.81GIPDD1850 pKa = 3.82HH1851 pKa = 7.37RR1852 pKa = 11.84DD1853 pKa = 3.17QFPQDD1858 pKa = 3.76PNASADD1864 pKa = 3.56GDD1866 pKa = 3.94GDD1868 pKa = 5.02GIPDD1872 pKa = 4.54SQDD1875 pKa = 2.67TDD1877 pKa = 3.55KK1878 pKa = 11.79NNDD1881 pKa = 3.52GFPDD1885 pKa = 3.63DD1886 pKa = 4.22QIIVSSVVTPNQPGIEE1902 pKa = 4.25EE1903 pKa = 4.03TWKK1906 pKa = 10.68VINIEE1911 pKa = 4.32DD1912 pKa = 3.69YY1913 pKa = 9.46PYY1915 pKa = 10.48TSVRR1919 pKa = 11.84VYY1921 pKa = 10.88APDD1924 pKa = 3.62GSRR1927 pKa = 11.84VFQSINYY1934 pKa = 8.31KK1935 pKa = 10.36NDD1937 pKa = 2.62WRR1939 pKa = 11.84GTNIRR1944 pKa = 11.84TGRR1947 pKa = 11.84PLPTGPYY1954 pKa = 8.88YY1955 pKa = 10.97YY1956 pKa = 10.11RR1957 pKa = 11.84IEE1959 pKa = 4.29LGGTSGEE1966 pKa = 4.78IIDD1969 pKa = 3.52GWLYY1973 pKa = 10.67IFNN1976 pKa = 4.53
Molecular weight: 210.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U2RXD9|A0A2U2RXD9_9BACT Acetolactate synthase OS=Bacteroidetes bacterium SCGC AAA795-G10 OX=2175803 GN=ilvB PE=3 SV=1
MM1 pKa = 7.41KK2 pKa = 10.33KK3 pKa = 8.37STKK6 pKa = 9.95VRR8 pKa = 11.84IFFEE12 pKa = 4.41RR13 pKa = 11.84NGFAVCSRR21 pKa = 11.84LAEE24 pKa = 4.01ILGMRR29 pKa = 11.84VKK31 pKa = 10.36SVRR34 pKa = 11.84LFFIYY39 pKa = 10.45ASFTTLVGGFVLYY52 pKa = 10.79LILALWLKK60 pKa = 10.61LNNLIYY66 pKa = 10.12TKK68 pKa = 10.18RR69 pKa = 11.84ASVFDD74 pKa = 3.9LL75 pKa = 4.15
MM1 pKa = 7.41KK2 pKa = 10.33KK3 pKa = 8.37STKK6 pKa = 9.95VRR8 pKa = 11.84IFFEE12 pKa = 4.41RR13 pKa = 11.84NGFAVCSRR21 pKa = 11.84LAEE24 pKa = 4.01ILGMRR29 pKa = 11.84VKK31 pKa = 10.36SVRR34 pKa = 11.84LFFIYY39 pKa = 10.45ASFTTLVGGFVLYY52 pKa = 10.79LILALWLKK60 pKa = 10.61LNNLIYY66 pKa = 10.12TKK68 pKa = 10.18RR69 pKa = 11.84ASVFDD74 pKa = 3.9LL75 pKa = 4.15
Molecular weight: 8.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
599355 |
33 |
5491 |
339.2 |
38.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.139 ± 0.057 | 0.783 ± 0.017 |
5.445 ± 0.107 | 6.07 ± 0.066 |
5.577 ± 0.062 | 6.498 ± 0.072 |
1.615 ± 0.031 | 8.955 ± 0.07 |
8.531 ± 0.116 | 9.465 ± 0.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.012 ± 0.027 | 6.737 ± 0.066 |
3.513 ± 0.032 | 3.096 ± 0.03 |
3.493 ± 0.039 | 7.506 ± 0.074 |
5.065 ± 0.088 | 5.436 ± 0.04 |
1.145 ± 0.023 | 3.918 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
