
Human papillomavirus 95
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 1
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
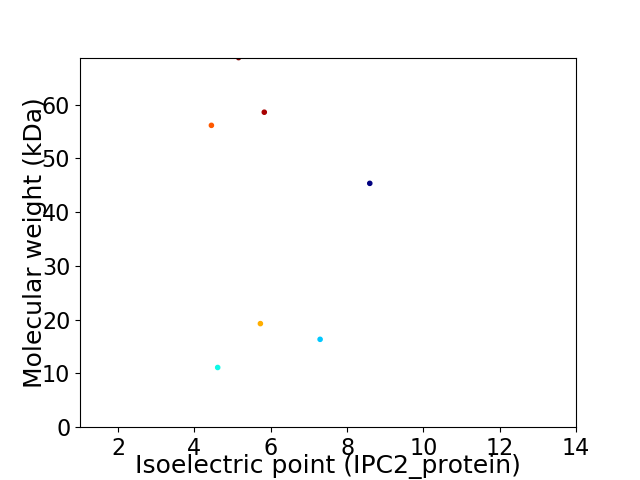
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q705D9|Q705D9_9PAPI Protein E6 OS=Human papillomavirus 95 OX=260716 GN=E6 PE=3 SV=1
MM1 pKa = 7.66RR2 pKa = 11.84GAAPSIADD10 pKa = 3.08VDD12 pKa = 4.75LNLHH16 pKa = 5.7EE17 pKa = 4.95LVVPANLLSDD27 pKa = 4.06EE28 pKa = 4.19VLQLSEE34 pKa = 4.59EE35 pKa = 4.11EE36 pKa = 4.09DD37 pKa = 3.57EE38 pKa = 4.34EE39 pKa = 5.32RR40 pKa = 11.84EE41 pKa = 4.1EE42 pKa = 3.9EE43 pKa = 4.21LLPFRR48 pKa = 11.84IDD50 pKa = 3.36TCCYY54 pKa = 8.45NCEE57 pKa = 3.98ANVRR61 pKa = 11.84ITLYY65 pKa = 10.87AVAFGLRR72 pKa = 11.84VVEE75 pKa = 4.13QLLLEE80 pKa = 4.43GKK82 pKa = 10.39VIFCCVGCARR92 pKa = 11.84NHH94 pKa = 5.62SRR96 pKa = 11.84NGRR99 pKa = 3.51
MM1 pKa = 7.66RR2 pKa = 11.84GAAPSIADD10 pKa = 3.08VDD12 pKa = 4.75LNLHH16 pKa = 5.7EE17 pKa = 4.95LVVPANLLSDD27 pKa = 4.06EE28 pKa = 4.19VLQLSEE34 pKa = 4.59EE35 pKa = 4.11EE36 pKa = 4.09DD37 pKa = 3.57EE38 pKa = 4.34EE39 pKa = 5.32RR40 pKa = 11.84EE41 pKa = 4.1EE42 pKa = 3.9EE43 pKa = 4.21LLPFRR48 pKa = 11.84IDD50 pKa = 3.36TCCYY54 pKa = 8.45NCEE57 pKa = 3.98ANVRR61 pKa = 11.84ITLYY65 pKa = 10.87AVAFGLRR72 pKa = 11.84VVEE75 pKa = 4.13QLLLEE80 pKa = 4.43GKK82 pKa = 10.39VIFCCVGCARR92 pKa = 11.84NHH94 pKa = 5.62SRR96 pKa = 11.84NGRR99 pKa = 3.51
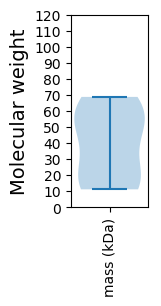
Molecular weight: 11.11 kDa
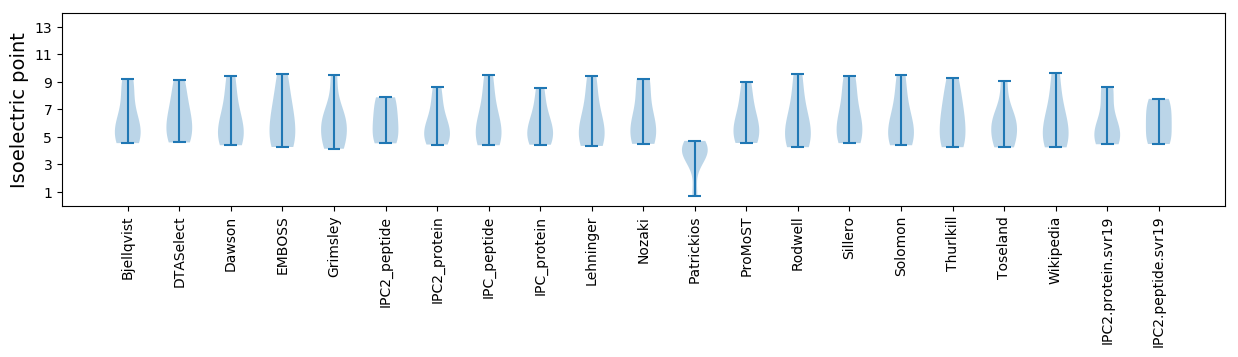
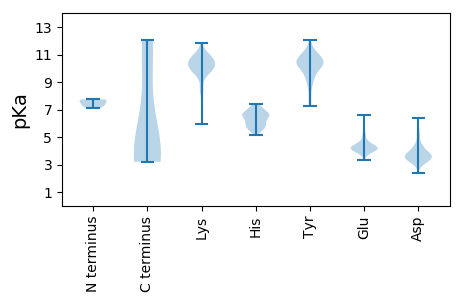
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q705D7|Q705D7_9PAPI Replication protein E1 OS=Human papillomavirus 95 OX=260716 GN=E1 PE=3 SV=1
MM1 pKa = 7.49EE2 pKa = 4.93SLVARR7 pKa = 11.84FDD9 pKa = 3.76ALQEE13 pKa = 4.31EE14 pKa = 5.26ILTHH18 pKa = 6.12IEE20 pKa = 4.05SGNTTLEE27 pKa = 3.98SQIKK31 pKa = 8.43YY32 pKa = 8.62WEE34 pKa = 4.07NVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 4.08NAIMHH44 pKa = 6.27YY45 pKa = 10.3ARR47 pKa = 11.84KK48 pKa = 9.64QGLTKK53 pKa = 10.71LGLQPLPSLLASEE66 pKa = 5.1YY67 pKa = 9.95NAKK70 pKa = 9.87QAIQIQLTLLSLLKK84 pKa = 10.42SPYY87 pKa = 10.39ASEE90 pKa = 4.14PWTLPEE96 pKa = 4.76VSAEE100 pKa = 4.51LINTPPQNVLKK111 pKa = 10.8KK112 pKa = 10.1GGYY115 pKa = 9.85DD116 pKa = 3.23VTVWFDD122 pKa = 3.53DD123 pKa = 3.64DD124 pKa = 4.72RR125 pKa = 11.84NNTMVYY131 pKa = 9.33TNWTALYY138 pKa = 9.14YY139 pKa = 10.38QDD141 pKa = 5.8ANEE144 pKa = 3.99IWHH147 pKa = 5.82KK148 pKa = 11.04VKK150 pKa = 11.28GEE152 pKa = 3.56VDD154 pKa = 3.54YY155 pKa = 11.87NGLFFTDD162 pKa = 3.49HH163 pKa = 5.87TGEE166 pKa = 3.88RR167 pKa = 11.84AYY169 pKa = 9.63FTLFSTDD176 pKa = 3.02AEE178 pKa = 4.45RR179 pKa = 11.84YY180 pKa = 8.4SQTGLWTVHH189 pKa = 6.5FKK191 pKa = 9.38TQVISSSVVSSTNPSSFDD209 pKa = 3.89FEE211 pKa = 4.35EE212 pKa = 4.16QLPGPSTSNTKK223 pKa = 7.1TTKK226 pKa = 8.45QTSPRR231 pKa = 11.84GRR233 pKa = 11.84GSQSRR238 pKa = 11.84EE239 pKa = 3.92LQPSSTTSPEE249 pKa = 4.12GKK251 pKa = 9.4GLRR254 pKa = 11.84VRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84GQGEE263 pKa = 4.33SGSGTRR269 pKa = 11.84EE270 pKa = 3.88TPSKK274 pKa = 10.16RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84GGGGRR282 pKa = 11.84GEE284 pKa = 4.51KK285 pKa = 10.35EE286 pKa = 4.09FGSAPTPSEE295 pKa = 3.65VGSRR299 pKa = 11.84HH300 pKa = 5.18RR301 pKa = 11.84QVEE304 pKa = 4.54TKK306 pKa = 10.28GLSRR310 pKa = 11.84LGQLQADD317 pKa = 3.78ARR319 pKa = 11.84DD320 pKa = 3.75PPMIMLKK327 pKa = 10.41GHH329 pKa = 6.64ANSLKK334 pKa = 8.57CWRR337 pKa = 11.84YY338 pKa = 9.8RR339 pKa = 11.84KK340 pKa = 8.39LTSNSCGFLYY350 pKa = 10.27MSTVWNWVGDD360 pKa = 3.62SSEE363 pKa = 3.91NHH365 pKa = 5.49SRR367 pKa = 11.84MLIAFTSTDD376 pKa = 3.11QRR378 pKa = 11.84DD379 pKa = 4.26YY380 pKa = 10.92FVKK383 pKa = 9.99HH384 pKa = 5.95HH385 pKa = 6.63FFPRR389 pKa = 11.84QCTYY393 pKa = 10.89TFGSLNSLL401 pKa = 3.86
MM1 pKa = 7.49EE2 pKa = 4.93SLVARR7 pKa = 11.84FDD9 pKa = 3.76ALQEE13 pKa = 4.31EE14 pKa = 5.26ILTHH18 pKa = 6.12IEE20 pKa = 4.05SGNTTLEE27 pKa = 3.98SQIKK31 pKa = 8.43YY32 pKa = 8.62WEE34 pKa = 4.07NVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 4.08NAIMHH44 pKa = 6.27YY45 pKa = 10.3ARR47 pKa = 11.84KK48 pKa = 9.64QGLTKK53 pKa = 10.71LGLQPLPSLLASEE66 pKa = 5.1YY67 pKa = 9.95NAKK70 pKa = 9.87QAIQIQLTLLSLLKK84 pKa = 10.42SPYY87 pKa = 10.39ASEE90 pKa = 4.14PWTLPEE96 pKa = 4.76VSAEE100 pKa = 4.51LINTPPQNVLKK111 pKa = 10.8KK112 pKa = 10.1GGYY115 pKa = 9.85DD116 pKa = 3.23VTVWFDD122 pKa = 3.53DD123 pKa = 3.64DD124 pKa = 4.72RR125 pKa = 11.84NNTMVYY131 pKa = 9.33TNWTALYY138 pKa = 9.14YY139 pKa = 10.38QDD141 pKa = 5.8ANEE144 pKa = 3.99IWHH147 pKa = 5.82KK148 pKa = 11.04VKK150 pKa = 11.28GEE152 pKa = 3.56VDD154 pKa = 3.54YY155 pKa = 11.87NGLFFTDD162 pKa = 3.49HH163 pKa = 5.87TGEE166 pKa = 3.88RR167 pKa = 11.84AYY169 pKa = 9.63FTLFSTDD176 pKa = 3.02AEE178 pKa = 4.45RR179 pKa = 11.84YY180 pKa = 8.4SQTGLWTVHH189 pKa = 6.5FKK191 pKa = 9.38TQVISSSVVSSTNPSSFDD209 pKa = 3.89FEE211 pKa = 4.35EE212 pKa = 4.16QLPGPSTSNTKK223 pKa = 7.1TTKK226 pKa = 8.45QTSPRR231 pKa = 11.84GRR233 pKa = 11.84GSQSRR238 pKa = 11.84EE239 pKa = 3.92LQPSSTTSPEE249 pKa = 4.12GKK251 pKa = 9.4GLRR254 pKa = 11.84VRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84GQGEE263 pKa = 4.33SGSGTRR269 pKa = 11.84EE270 pKa = 3.88TPSKK274 pKa = 10.16RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84GGGGRR282 pKa = 11.84GEE284 pKa = 4.51KK285 pKa = 10.35EE286 pKa = 4.09FGSAPTPSEE295 pKa = 3.65VGSRR299 pKa = 11.84HH300 pKa = 5.18RR301 pKa = 11.84QVEE304 pKa = 4.54TKK306 pKa = 10.28GLSRR310 pKa = 11.84LGQLQADD317 pKa = 3.78ARR319 pKa = 11.84DD320 pKa = 3.75PPMIMLKK327 pKa = 10.41GHH329 pKa = 6.64ANSLKK334 pKa = 8.57CWRR337 pKa = 11.84YY338 pKa = 9.8RR339 pKa = 11.84KK340 pKa = 8.39LTSNSCGFLYY350 pKa = 10.27MSTVWNWVGDD360 pKa = 3.62SSEE363 pKa = 3.91NHH365 pKa = 5.49SRR367 pKa = 11.84MLIAFTSTDD376 pKa = 3.11QRR378 pKa = 11.84DD379 pKa = 4.26YY380 pKa = 10.92FVKK383 pKa = 9.99HH384 pKa = 5.95HH385 pKa = 6.63FFPRR389 pKa = 11.84QCTYY393 pKa = 10.89TFGSLNSLL401 pKa = 3.86
Molecular weight: 45.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2447 |
99 |
601 |
349.6 |
39.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.517 ± 0.414 | 2.207 ± 0.718 |
6.457 ± 0.646 | 6.293 ± 0.516 |
4.373 ± 0.574 | 6.416 ± 0.833 |
1.839 ± 0.27 | 5.272 ± 0.859 |
4.863 ± 0.906 | 9.44 ± 0.653 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.389 ± 0.324 | 4.373 ± 0.51 |
6.089 ± 1.047 | 4.863 ± 0.263 |
5.926 ± 0.534 | 7.928 ± 0.844 |
6.293 ± 0.857 | 5.517 ± 0.515 |
1.226 ± 0.36 | 3.719 ± 0.381 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
