
Mesta yellow vein mosaic Bahraich virus-[India:Bahraich:2007]
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; Mesta yellow vein mosaic Bahraich virus
Average proteome isoelectric point is 7.97
Get precalculated fractions of proteins
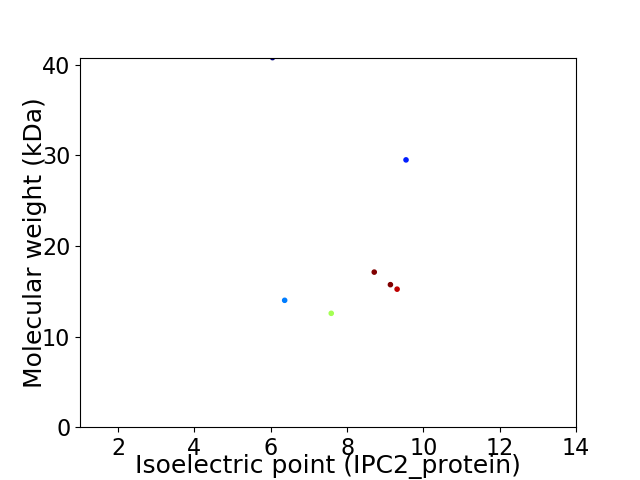
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1PAJ0|B1PAJ0_9GEMI AC4 OS=Mesta yellow vein mosaic Bahraich virus-[India:Bahraich:2007] OX=508749 GN=AC4 PE=3 SV=1
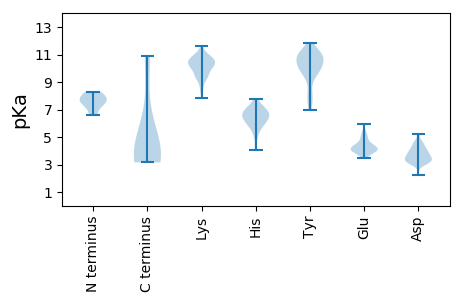
MM1 pKa = 7.74PPRR4 pKa = 11.84RR5 pKa = 11.84NGIYY9 pKa = 10.25SKK11 pKa = 10.68DD12 pKa = 3.68YY13 pKa = 9.69FVTYY17 pKa = 8.32PKK19 pKa = 10.65CSLTKK24 pKa = 10.69EE25 pKa = 4.12EE26 pKa = 5.36ALSQLLNLQTPTSKK40 pKa = 10.57KK41 pKa = 9.41YY42 pKa = 10.62IRR44 pKa = 11.84ICRR47 pKa = 11.84EE48 pKa = 3.51LHH50 pKa = 6.09EE51 pKa = 5.96DD52 pKa = 3.11GTPRR56 pKa = 11.84VRR58 pKa = 11.84VLIQFEE64 pKa = 4.97GKK66 pKa = 10.06FKK68 pKa = 9.6CQNMRR73 pKa = 11.84FFDD76 pKa = 4.16LVSPSWSAHH85 pKa = 4.35FRR87 pKa = 11.84PNIQGAKK94 pKa = 9.58SSSDD98 pKa = 3.06VKK100 pKa = 11.23SYY102 pKa = 10.8IEE104 pKa = 4.18KK105 pKa = 10.92DD106 pKa = 3.16GDD108 pKa = 3.65ILDD111 pKa = 3.83WGQFQIDD118 pKa = 3.44GRR120 pKa = 11.84SARR123 pKa = 11.84GGQQTANDD131 pKa = 4.05AYY133 pKa = 10.69AAALNAGSKK142 pKa = 10.26SEE144 pKa = 4.31AIRR147 pKa = 11.84VIKK150 pKa = 10.15EE151 pKa = 3.57LAPKK155 pKa = 10.6DD156 pKa = 3.7FVLQFHH162 pKa = 6.7NLNANLNRR170 pKa = 11.84IFQEE174 pKa = 4.12PPAPYY179 pKa = 9.91VSPFSSSSFDD189 pKa = 3.41QVPEE193 pKa = 4.03EE194 pKa = 4.1LEE196 pKa = 3.86EE197 pKa = 4.02WASEE201 pKa = 4.17NVVDD205 pKa = 5.24AAARR209 pKa = 11.84PLRR212 pKa = 11.84PQSIVIEE219 pKa = 4.44GDD221 pKa = 3.12SRR223 pKa = 11.84TGKK226 pKa = 8.52TMWARR231 pKa = 11.84SLGPHH236 pKa = 6.71NYY238 pKa = 10.18LCGHH242 pKa = 7.38LDD244 pKa = 4.18LSPKK248 pKa = 10.15VYY250 pKa = 11.07SNDD253 pKa = 2.25AWYY256 pKa = 10.92NVIDD260 pKa = 5.2DD261 pKa = 4.41VDD263 pKa = 3.61PHH265 pKa = 5.85YY266 pKa = 11.0LKK268 pKa = 10.7HH269 pKa = 6.2FKK271 pKa = 10.7EE272 pKa = 4.46FMGAQRR278 pKa = 11.84DD279 pKa = 3.81WQSNTKK285 pKa = 9.6YY286 pKa = 10.57GKK288 pKa = 9.12PVQIKK293 pKa = 10.41GGIPTIFLCNPGPNASYY310 pKa = 11.2KK311 pKa = 10.7EE312 pKa = 3.97FLDD315 pKa = 3.54EE316 pKa = 5.43DD317 pKa = 4.1KK318 pKa = 11.59NSALKK323 pKa = 10.43SWALKK328 pKa = 9.99NATFVFLTQPLYY340 pKa = 11.2SGTNQSSTQGSEE352 pKa = 3.74EE353 pKa = 3.96AQQEE357 pKa = 4.53EE358 pKa = 4.95TSGPP362 pKa = 3.61
MM1 pKa = 7.74PPRR4 pKa = 11.84RR5 pKa = 11.84NGIYY9 pKa = 10.25SKK11 pKa = 10.68DD12 pKa = 3.68YY13 pKa = 9.69FVTYY17 pKa = 8.32PKK19 pKa = 10.65CSLTKK24 pKa = 10.69EE25 pKa = 4.12EE26 pKa = 5.36ALSQLLNLQTPTSKK40 pKa = 10.57KK41 pKa = 9.41YY42 pKa = 10.62IRR44 pKa = 11.84ICRR47 pKa = 11.84EE48 pKa = 3.51LHH50 pKa = 6.09EE51 pKa = 5.96DD52 pKa = 3.11GTPRR56 pKa = 11.84VRR58 pKa = 11.84VLIQFEE64 pKa = 4.97GKK66 pKa = 10.06FKK68 pKa = 9.6CQNMRR73 pKa = 11.84FFDD76 pKa = 4.16LVSPSWSAHH85 pKa = 4.35FRR87 pKa = 11.84PNIQGAKK94 pKa = 9.58SSSDD98 pKa = 3.06VKK100 pKa = 11.23SYY102 pKa = 10.8IEE104 pKa = 4.18KK105 pKa = 10.92DD106 pKa = 3.16GDD108 pKa = 3.65ILDD111 pKa = 3.83WGQFQIDD118 pKa = 3.44GRR120 pKa = 11.84SARR123 pKa = 11.84GGQQTANDD131 pKa = 4.05AYY133 pKa = 10.69AAALNAGSKK142 pKa = 10.26SEE144 pKa = 4.31AIRR147 pKa = 11.84VIKK150 pKa = 10.15EE151 pKa = 3.57LAPKK155 pKa = 10.6DD156 pKa = 3.7FVLQFHH162 pKa = 6.7NLNANLNRR170 pKa = 11.84IFQEE174 pKa = 4.12PPAPYY179 pKa = 9.91VSPFSSSSFDD189 pKa = 3.41QVPEE193 pKa = 4.03EE194 pKa = 4.1LEE196 pKa = 3.86EE197 pKa = 4.02WASEE201 pKa = 4.17NVVDD205 pKa = 5.24AAARR209 pKa = 11.84PLRR212 pKa = 11.84PQSIVIEE219 pKa = 4.44GDD221 pKa = 3.12SRR223 pKa = 11.84TGKK226 pKa = 8.52TMWARR231 pKa = 11.84SLGPHH236 pKa = 6.71NYY238 pKa = 10.18LCGHH242 pKa = 7.38LDD244 pKa = 4.18LSPKK248 pKa = 10.15VYY250 pKa = 11.07SNDD253 pKa = 2.25AWYY256 pKa = 10.92NVIDD260 pKa = 5.2DD261 pKa = 4.41VDD263 pKa = 3.61PHH265 pKa = 5.85YY266 pKa = 11.0LKK268 pKa = 10.7HH269 pKa = 6.2FKK271 pKa = 10.7EE272 pKa = 4.46FMGAQRR278 pKa = 11.84DD279 pKa = 3.81WQSNTKK285 pKa = 9.6YY286 pKa = 10.57GKK288 pKa = 9.12PVQIKK293 pKa = 10.41GGIPTIFLCNPGPNASYY310 pKa = 11.2KK311 pKa = 10.7EE312 pKa = 3.97FLDD315 pKa = 3.54EE316 pKa = 5.43DD317 pKa = 4.1KK318 pKa = 11.59NSALKK323 pKa = 10.43SWALKK328 pKa = 9.99NATFVFLTQPLYY340 pKa = 11.2SGTNQSSTQGSEE352 pKa = 3.74EE353 pKa = 3.96AQQEE357 pKa = 4.53EE358 pKa = 4.95TSGPP362 pKa = 3.61
Molecular weight: 40.79 kDa
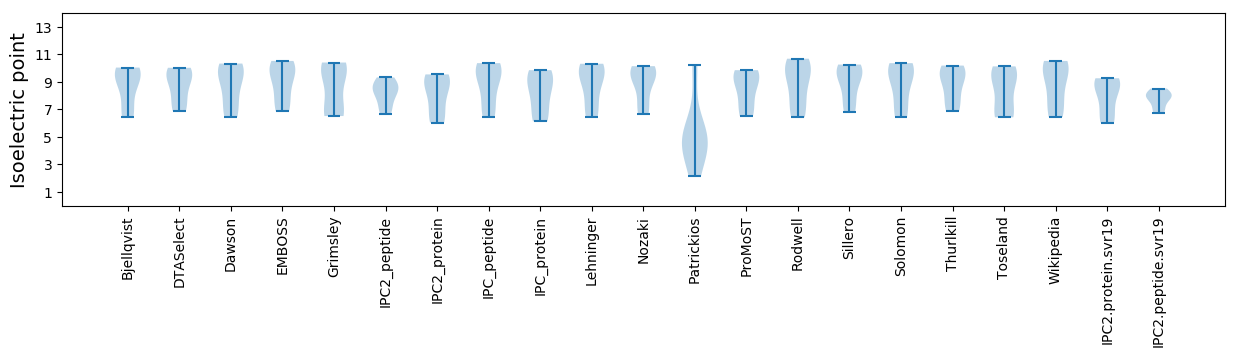
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B1PAI7|B1PAI7_9GEMI Replication enhancer OS=Mesta yellow vein mosaic Bahraich virus-[India:Bahraich:2007] OX=508749 GN=AC3 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 10.21RR4 pKa = 11.84AADD7 pKa = 3.55IVISTPASKK16 pKa = 10.29VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFGSPYY27 pKa = 7.94TSRR30 pKa = 11.84VAAPIVRR37 pKa = 11.84VTKK40 pKa = 9.91QQAWTNRR47 pKa = 11.84PMNRR51 pKa = 11.84KK52 pKa = 7.8PRR54 pKa = 11.84MYY56 pKa = 10.68RR57 pKa = 11.84MYY59 pKa = 10.44RR60 pKa = 11.84SPDD63 pKa = 3.21VPRR66 pKa = 11.84GCEE69 pKa = 4.34GPCKK73 pKa = 9.9VQSFVSTPDD82 pKa = 3.06VVHH85 pKa = 6.88IGKK88 pKa = 9.48VMCISDD94 pKa = 3.53VTRR97 pKa = 11.84GVGLTHH103 pKa = 7.8RR104 pKa = 11.84IGKK107 pKa = 8.54RR108 pKa = 11.84FCVKK112 pKa = 9.96SVYY115 pKa = 10.52VLGKK119 pKa = 9.41IWMDD123 pKa = 3.35EE124 pKa = 4.04NIKK127 pKa = 9.32TKK129 pKa = 10.6NHH131 pKa = 5.78TNSVMFFLVRR141 pKa = 11.84DD142 pKa = 3.84RR143 pKa = 11.84RR144 pKa = 11.84PVDD147 pKa = 3.13KK148 pKa = 10.28PQDD151 pKa = 3.56FGEE154 pKa = 4.33VFNMFDD160 pKa = 4.29NEE162 pKa = 4.11PSTATVKK169 pKa = 10.57NSHH172 pKa = 6.59RR173 pKa = 11.84DD174 pKa = 3.15RR175 pKa = 11.84YY176 pKa = 9.17QVLRR180 pKa = 11.84KK181 pKa = 8.43WHH183 pKa = 5.58ATVTGGQYY191 pKa = 11.09ASNEE195 pKa = 3.8HH196 pKa = 6.49ALVKK200 pKa = 10.69KK201 pKa = 9.38FVRR204 pKa = 11.84VNNYY208 pKa = 7.82VVYY211 pKa = 9.96NQQEE215 pKa = 3.8AGKK218 pKa = 10.1YY219 pKa = 8.23EE220 pKa = 4.02NHH222 pKa = 6.47TEE224 pKa = 3.99NALMLYY230 pKa = 7.52MACTHH235 pKa = 7.06ASNPVYY241 pKa = 10.61APLKK245 pKa = 9.09IRR247 pKa = 11.84IYY249 pKa = 10.62FYY251 pKa = 11.34DD252 pKa = 3.32SVTNN256 pKa = 3.98
MM1 pKa = 7.69SKK3 pKa = 10.21RR4 pKa = 11.84AADD7 pKa = 3.55IVISTPASKK16 pKa = 10.29VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFGSPYY27 pKa = 7.94TSRR30 pKa = 11.84VAAPIVRR37 pKa = 11.84VTKK40 pKa = 9.91QQAWTNRR47 pKa = 11.84PMNRR51 pKa = 11.84KK52 pKa = 7.8PRR54 pKa = 11.84MYY56 pKa = 10.68RR57 pKa = 11.84MYY59 pKa = 10.44RR60 pKa = 11.84SPDD63 pKa = 3.21VPRR66 pKa = 11.84GCEE69 pKa = 4.34GPCKK73 pKa = 9.9VQSFVSTPDD82 pKa = 3.06VVHH85 pKa = 6.88IGKK88 pKa = 9.48VMCISDD94 pKa = 3.53VTRR97 pKa = 11.84GVGLTHH103 pKa = 7.8RR104 pKa = 11.84IGKK107 pKa = 8.54RR108 pKa = 11.84FCVKK112 pKa = 9.96SVYY115 pKa = 10.52VLGKK119 pKa = 9.41IWMDD123 pKa = 3.35EE124 pKa = 4.04NIKK127 pKa = 9.32TKK129 pKa = 10.6NHH131 pKa = 5.78TNSVMFFLVRR141 pKa = 11.84DD142 pKa = 3.84RR143 pKa = 11.84RR144 pKa = 11.84PVDD147 pKa = 3.13KK148 pKa = 10.28PQDD151 pKa = 3.56FGEE154 pKa = 4.33VFNMFDD160 pKa = 4.29NEE162 pKa = 4.11PSTATVKK169 pKa = 10.57NSHH172 pKa = 6.59RR173 pKa = 11.84DD174 pKa = 3.15RR175 pKa = 11.84YY176 pKa = 9.17QVLRR180 pKa = 11.84KK181 pKa = 8.43WHH183 pKa = 5.58ATVTGGQYY191 pKa = 11.09ASNEE195 pKa = 3.8HH196 pKa = 6.49ALVKK200 pKa = 10.69KK201 pKa = 9.38FVRR204 pKa = 11.84VNNYY208 pKa = 7.82VVYY211 pKa = 9.96NQQEE215 pKa = 3.8AGKK218 pKa = 10.1YY219 pKa = 8.23EE220 pKa = 4.02NHH222 pKa = 6.47TEE224 pKa = 3.99NALMLYY230 pKa = 7.52MACTHH235 pKa = 7.06ASNPVYY241 pKa = 10.61APLKK245 pKa = 9.09IRR247 pKa = 11.84IYY249 pKa = 10.62FYY251 pKa = 11.34DD252 pKa = 3.32SVTNN256 pKa = 3.98
Molecular weight: 29.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1269 |
112 |
362 |
181.3 |
20.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.886 ± 0.821 | 2.522 ± 0.665 |
4.728 ± 0.372 | 4.177 ± 0.688 |
3.94 ± 0.536 | 5.28 ± 0.32 |
3.625 ± 0.688 | 4.728 ± 0.601 |
5.201 ± 0.791 | 7.486 ± 1.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.049 ± 0.469 | 6.462 ± 0.722 |
5.989 ± 0.41 | 4.886 ± 0.773 |
7.171 ± 0.797 | 8.905 ± 1.279 |
6.068 ± 0.969 | 6.304 ± 1.347 |
1.497 ± 0.142 | 4.098 ± 0.408 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
