
Cryobacterium sp. Hh39
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
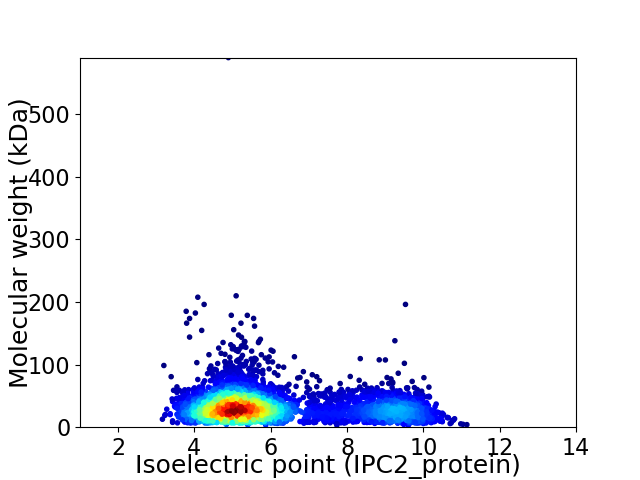
Virtual 2D-PAGE plot for 4157 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A5MGS3|A0A3A5MGS3_9MICO 4-hydroxy-4-methyl-2-oxoglutarate aldolase OS=Cryobacterium sp. Hh39 OX=995039 GN=D6T64_07085 PE=3 SV=1
MM1 pKa = 7.43HH2 pKa = 7.25KK3 pKa = 10.21RR4 pKa = 11.84YY5 pKa = 8.74FAAPALVAAAALALSGCASTPASSSAADD33 pKa = 3.29GDD35 pKa = 4.11LRR37 pKa = 11.84VVATTTQIADD47 pKa = 3.79FTRR50 pKa = 11.84SVVGDD55 pKa = 3.55AAGVSITQLVQPNQSLHH72 pKa = 7.06SYY74 pKa = 10.25DD75 pKa = 4.1PSAADD80 pKa = 3.61LTALGAADD88 pKa = 3.34VLVINGVDD96 pKa = 3.6LEE98 pKa = 4.33EE99 pKa = 4.43WLDD102 pKa = 3.8DD103 pKa = 5.27AIDD106 pKa = 3.65ASGFDD111 pKa = 3.98GVTIDD116 pKa = 3.82SDD118 pKa = 3.57EE119 pKa = 5.13GIDD122 pKa = 3.69IIEE125 pKa = 4.42SEE127 pKa = 4.28VDD129 pKa = 3.17EE130 pKa = 4.79HH131 pKa = 9.31AEE133 pKa = 3.94DD134 pKa = 4.43HH135 pKa = 7.19ADD137 pKa = 3.61EE138 pKa = 4.45TDD140 pKa = 3.47EE141 pKa = 3.89EE142 pKa = 4.54HH143 pKa = 7.0AAEE146 pKa = 4.1EE147 pKa = 3.92AAAAAEE153 pKa = 4.3EE154 pKa = 4.58AEE156 pKa = 4.51HH157 pKa = 7.56ADD159 pKa = 3.59EE160 pKa = 4.73TEE162 pKa = 4.16EE163 pKa = 3.79EE164 pKa = 4.37HH165 pKa = 7.27AAEE168 pKa = 5.17DD169 pKa = 4.35DD170 pKa = 4.14GEE172 pKa = 4.23HH173 pKa = 6.2AHH175 pKa = 7.55DD176 pKa = 5.43GGDD179 pKa = 3.36PHH181 pKa = 8.45IWTDD185 pKa = 3.21VNNAMKK191 pKa = 10.03MVGTIAGGLEE201 pKa = 3.98EE202 pKa = 5.29ALSADD207 pKa = 4.04SADD210 pKa = 3.18TAAAIEE216 pKa = 4.39ANATAYY222 pKa = 10.11SQQLSEE228 pKa = 4.3LDD230 pKa = 2.98SWIRR234 pKa = 11.84TNVDD238 pKa = 3.23TVPAAEE244 pKa = 4.28RR245 pKa = 11.84LLVSNHH251 pKa = 6.75DD252 pKa = 3.54ALGYY256 pKa = 7.99YY257 pKa = 7.99TAAYY261 pKa = 9.24GITYY265 pKa = 9.47VGAVIPSFDD274 pKa = 4.64DD275 pKa = 3.41NAEE278 pKa = 4.05PSAAAIDD285 pKa = 4.03EE286 pKa = 4.27LVAAITATGVQAVFSEE302 pKa = 4.5ASLNPKK308 pKa = 8.6TADD311 pKa = 3.72TIASEE316 pKa = 4.6ANVTVYY322 pKa = 10.76SGDD325 pKa = 3.39DD326 pKa = 3.27ALYY329 pKa = 10.66VDD331 pKa = 4.86SLGPDD336 pKa = 3.9GSAGATYY343 pKa = 10.31IAAQLHH349 pKa = 5.33NTTAILEE356 pKa = 4.46SWGVTPTDD364 pKa = 3.4VPADD368 pKa = 3.74LEE370 pKa = 4.37KK371 pKa = 11.14
MM1 pKa = 7.43HH2 pKa = 7.25KK3 pKa = 10.21RR4 pKa = 11.84YY5 pKa = 8.74FAAPALVAAAALALSGCASTPASSSAADD33 pKa = 3.29GDD35 pKa = 4.11LRR37 pKa = 11.84VVATTTQIADD47 pKa = 3.79FTRR50 pKa = 11.84SVVGDD55 pKa = 3.55AAGVSITQLVQPNQSLHH72 pKa = 7.06SYY74 pKa = 10.25DD75 pKa = 4.1PSAADD80 pKa = 3.61LTALGAADD88 pKa = 3.34VLVINGVDD96 pKa = 3.6LEE98 pKa = 4.33EE99 pKa = 4.43WLDD102 pKa = 3.8DD103 pKa = 5.27AIDD106 pKa = 3.65ASGFDD111 pKa = 3.98GVTIDD116 pKa = 3.82SDD118 pKa = 3.57EE119 pKa = 5.13GIDD122 pKa = 3.69IIEE125 pKa = 4.42SEE127 pKa = 4.28VDD129 pKa = 3.17EE130 pKa = 4.79HH131 pKa = 9.31AEE133 pKa = 3.94DD134 pKa = 4.43HH135 pKa = 7.19ADD137 pKa = 3.61EE138 pKa = 4.45TDD140 pKa = 3.47EE141 pKa = 3.89EE142 pKa = 4.54HH143 pKa = 7.0AAEE146 pKa = 4.1EE147 pKa = 3.92AAAAAEE153 pKa = 4.3EE154 pKa = 4.58AEE156 pKa = 4.51HH157 pKa = 7.56ADD159 pKa = 3.59EE160 pKa = 4.73TEE162 pKa = 4.16EE163 pKa = 3.79EE164 pKa = 4.37HH165 pKa = 7.27AAEE168 pKa = 5.17DD169 pKa = 4.35DD170 pKa = 4.14GEE172 pKa = 4.23HH173 pKa = 6.2AHH175 pKa = 7.55DD176 pKa = 5.43GGDD179 pKa = 3.36PHH181 pKa = 8.45IWTDD185 pKa = 3.21VNNAMKK191 pKa = 10.03MVGTIAGGLEE201 pKa = 3.98EE202 pKa = 5.29ALSADD207 pKa = 4.04SADD210 pKa = 3.18TAAAIEE216 pKa = 4.39ANATAYY222 pKa = 10.11SQQLSEE228 pKa = 4.3LDD230 pKa = 2.98SWIRR234 pKa = 11.84TNVDD238 pKa = 3.23TVPAAEE244 pKa = 4.28RR245 pKa = 11.84LLVSNHH251 pKa = 6.75DD252 pKa = 3.54ALGYY256 pKa = 7.99YY257 pKa = 7.99TAAYY261 pKa = 9.24GITYY265 pKa = 9.47VGAVIPSFDD274 pKa = 4.64DD275 pKa = 3.41NAEE278 pKa = 4.05PSAAAIDD285 pKa = 4.03EE286 pKa = 4.27LVAAITATGVQAVFSEE302 pKa = 4.5ASLNPKK308 pKa = 8.6TADD311 pKa = 3.72TIASEE316 pKa = 4.6ANVTVYY322 pKa = 10.76SGDD325 pKa = 3.39DD326 pKa = 3.27ALYY329 pKa = 10.66VDD331 pKa = 4.86SLGPDD336 pKa = 3.9GSAGATYY343 pKa = 10.31IAAQLHH349 pKa = 5.33NTTAILEE356 pKa = 4.46SWGVTPTDD364 pKa = 3.4VPADD368 pKa = 3.74LEE370 pKa = 4.37KK371 pKa = 11.14
Molecular weight: 38.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A5MSW1|A0A3A5MSW1_9MICO Uncharacterized protein OS=Cryobacterium sp. Hh39 OX=995039 GN=D6T64_02815 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1328963 |
29 |
5497 |
319.7 |
34.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.315 ± 0.056 | 0.559 ± 0.01 |
5.848 ± 0.036 | 5.11 ± 0.034 |
3.223 ± 0.022 | 8.745 ± 0.032 |
1.982 ± 0.016 | 4.694 ± 0.033 |
2.032 ± 0.027 | 10.447 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.812 ± 0.015 | 2.357 ± 0.02 |
5.367 ± 0.031 | 2.923 ± 0.019 |
6.65 ± 0.041 | 6.025 ± 0.029 |
6.777 ± 0.038 | 8.736 ± 0.034 |
1.411 ± 0.016 | 1.988 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
