
Duganella ginsengisoli
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Duganella
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
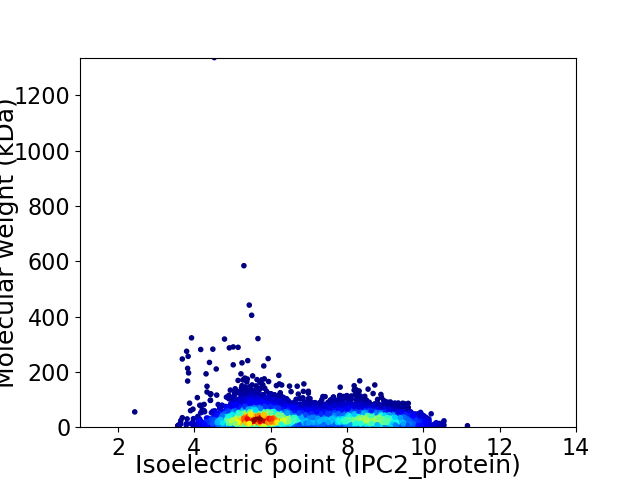
Virtual 2D-PAGE plot for 5918 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L6Q9A2|A0A6L6Q9A2_9BURK Uncharacterized protein OS=Duganella ginsengisoli OX=1462440 GN=GM668_29535 PE=4 SV=1
MM1 pKa = 7.86SDD3 pKa = 3.39LTAPSLKK10 pKa = 10.52SVTFTNSTANASAVNTSADD29 pKa = 3.09IVLTFSEE36 pKa = 4.78AMTSGTGLITISDD49 pKa = 4.49GATQTYY55 pKa = 8.45MGKK58 pKa = 10.2DD59 pKa = 2.98GTLRR63 pKa = 11.84TRR65 pKa = 11.84LVGATDD71 pKa = 3.26TRR73 pKa = 11.84TVDD76 pKa = 3.62ISDD79 pKa = 3.8TSQVTVSGDD88 pKa = 3.24TVTINLASDD97 pKa = 3.96LKK99 pKa = 10.78AGKK102 pKa = 9.89SYY104 pKa = 10.56SVQMGKK110 pKa = 10.57GVLLDD115 pKa = 3.78EE116 pKa = 5.65AGNAYY121 pKa = 10.31AGLSDD126 pKa = 3.5TTKK129 pKa = 11.15LKK131 pKa = 9.6FTTSAATLPTAVVDD145 pKa = 3.86STFTITDD152 pKa = 3.69TGSSASDD159 pKa = 4.47FITSSAVQTIAGKK172 pKa = 10.95VNGALLAGEE181 pKa = 4.42AVMVSLDD188 pKa = 4.74NGGTWAAATVNAADD202 pKa = 4.14STWSIGGTVTGSSTIIARR220 pKa = 11.84VVNAEE225 pKa = 4.09GNASAAARR233 pKa = 11.84HH234 pKa = 6.32AYY236 pKa = 9.06TFDD239 pKa = 3.25TDD241 pKa = 3.38APTVGSIAINDD252 pKa = 3.68TDD254 pKa = 4.12LVAGEE259 pKa = 4.32TATVTVTFNEE269 pKa = 4.36AVTGLDD275 pKa = 3.13VSDD278 pKa = 3.95FTVEE282 pKa = 4.03GGALSSLATADD293 pKa = 4.31NGKK296 pKa = 7.63TWTATLTPTKK306 pKa = 10.29GVSTTTGTVTLNAGSVADD324 pKa = 4.0LAGNSGPANSSSKK337 pKa = 9.78PFNYY341 pKa = 8.93STVPDD346 pKa = 4.14APTATVTSISMTDD359 pKa = 3.3TGSSATDD366 pKa = 4.37FITNSTSQTIKK377 pKa = 9.49GTYY380 pKa = 9.73SGTLAADD387 pKa = 4.28EE388 pKa = 4.75YY389 pKa = 10.97IQVSLDD395 pKa = 3.7NGSTWTKK402 pKa = 9.82ATVSGSNWEE411 pKa = 4.12VAGATISASNTLKK424 pKa = 10.79ARR426 pKa = 11.84VFNDD430 pKa = 3.19YY431 pKa = 10.2TYY433 pKa = 11.08SNPMAQAYY441 pKa = 7.86TLDD444 pKa = 3.55QTAPTATVTLDD455 pKa = 3.09KK456 pKa = 11.42AHH458 pKa = 6.61LKK460 pKa = 10.47KK461 pKa = 11.16GEE463 pKa = 4.17TATITITFSEE473 pKa = 4.79AVTGLTSADD482 pKa = 3.03IKK484 pKa = 11.62SNDD487 pKa = 3.65TVGTPVSTDD496 pKa = 3.05GGVTWTATLTPTNNGNGSVTLYY518 pKa = 10.76ASSVTDD524 pKa = 3.5LAGNSGPAADD534 pKa = 3.82VAATYY539 pKa = 10.78SSDD542 pKa = 3.35LTRR545 pKa = 11.84PTATISLADD554 pKa = 3.5SALTAGEE561 pKa = 4.36TTLVTITFSEE571 pKa = 4.44AVTEE575 pKa = 4.19FTNADD580 pKa = 3.6LTVGNGTLSTVTSTDD595 pKa = 3.2GGTTWTATLTPTASTTATTNVITLDD620 pKa = 3.47NTAVSDD626 pKa = 3.84LAGNTGIGSTSSANYY641 pKa = 9.31SVSTVRR647 pKa = 11.84PTATINIADD656 pKa = 4.02TALTAGEE663 pKa = 4.41TTTVTFTFSSAVTGFDD679 pKa = 3.3ASDD682 pKa = 3.29VTVANGTISTPTTTDD697 pKa = 3.13NITWTATLTPSASTEE712 pKa = 3.85DD713 pKa = 3.56TTNVITLANTGFHH726 pKa = 6.5NSAGNSGTGTTTSGNYY742 pKa = 8.72TVDD745 pKa = 3.35TARR748 pKa = 11.84PTATISLADD757 pKa = 3.49TALTVGEE764 pKa = 4.51TTTVTVKK771 pKa = 10.46FSEE774 pKa = 4.31AVTGFSASSLSVANGTISSLTSSDD798 pKa = 3.47NITWTGTLTPTASQEE813 pKa = 3.91DD814 pKa = 4.19STNVITLNNSGVSDD828 pKa = 3.61AAGNTGSGTTTSANYY843 pKa = 9.02TVDD846 pKa = 3.31TLRR849 pKa = 11.84PTATISLADD858 pKa = 3.5SALTAGEE865 pKa = 4.35TTTVSITFSEE875 pKa = 5.01VVTGLTTADD884 pKa = 3.35LTVANGTLTSLQSSDD899 pKa = 3.04GMNWTATLTPSASTDD914 pKa = 3.14DD915 pKa = 3.77TTNVITLDD923 pKa = 3.35NTGITDD929 pKa = 3.63ASGNAGSGTTTSANYY944 pKa = 8.53TVATVRR950 pKa = 11.84PTATIAVSDD959 pKa = 4.02TALKK963 pKa = 9.91TGDD966 pKa = 3.6TATVTITFSEE976 pKa = 4.55AVTGLTAADD985 pKa = 3.35FTVANGTLSTPTTTDD1000 pKa = 3.09NITWTATLTPTSSIEE1015 pKa = 3.9DD1016 pKa = 3.71TTNVITLDD1024 pKa = 3.54NTGVTDD1030 pKa = 3.43SDD1032 pKa = 4.3GNTGSGTTSSSNYY1045 pKa = 9.21TVDD1048 pKa = 3.26TKK1050 pKa = 11.59APTATIALSQPEE1062 pKa = 4.87LLTGEE1067 pKa = 4.36TSTVTITFNEE1077 pKa = 4.26AVTGLTLADD1086 pKa = 4.31FKK1088 pKa = 11.72YY1089 pKa = 10.16SGTMTAPVSTDD1100 pKa = 3.26GGTTWVATLTPSTEE1114 pKa = 4.09SSSGEE1119 pKa = 3.72VRR1121 pKa = 11.84LNTSSVTDD1129 pKa = 3.37SAGNSGPSSEE1139 pKa = 5.13AYY1141 pKa = 10.35ASFTYY1146 pKa = 10.35NAAPTATIATASITDD1161 pKa = 3.64TGTSGDD1167 pKa = 4.69FITSSTSQEE1176 pKa = 3.56ISGTLSAALGRR1187 pKa = 11.84GQYY1190 pKa = 9.81VQVSLDD1196 pKa = 3.59NGEE1199 pKa = 4.36SWTTATTIGTGWSLSGQTISHH1220 pKa = 6.52SSTLKK1225 pKa = 9.98VRR1227 pKa = 11.84ATNGSSTSTTYY1238 pKa = 10.48SHH1240 pKa = 7.41SYY1242 pKa = 9.6TLDD1245 pKa = 3.04QAAPTAPTITLSKK1258 pKa = 10.57DD1259 pKa = 3.08ALYY1262 pKa = 10.33TNQTATVTVVFDD1274 pKa = 3.44EE1275 pKa = 4.4AVYY1278 pKa = 10.95GLTASDD1284 pKa = 3.62FTISGGTLGTFSTSDD1299 pKa = 3.33NLTWTATLTPTATGSLALAASSVYY1323 pKa = 9.49DD1324 pKa = 3.3TAGNVGPTSASSSKK1338 pKa = 10.94SFTFTADD1345 pKa = 3.1TTGPTATLTLNDD1357 pKa = 3.44NTLGKK1362 pKa = 10.69GGTATLTVTLSEE1374 pKa = 4.6AVTDD1378 pKa = 3.99TPTASDD1384 pKa = 3.7FTVSGGSLGTFTSTDD1399 pKa = 3.07SGLTWTATLTPTDD1412 pKa = 3.54SAYY1415 pKa = 10.5GAGTISLSANAISDD1429 pKa = 3.5QSGNTGPSSAASLPFYY1445 pKa = 10.9YY1446 pKa = 9.36DD1447 pKa = 3.27TQALTLSSSIAFSTDD1462 pKa = 2.79SGEE1465 pKa = 4.31SGSDD1469 pKa = 3.68LITNTGTQTISGTYY1483 pKa = 9.53SGGTIYY1489 pKa = 10.04STMGDD1494 pKa = 3.82KK1495 pKa = 10.7IEE1497 pKa = 4.01VSTDD1501 pKa = 3.07NGSTWSEE1508 pKa = 3.83ATVSGQAWSISKK1520 pKa = 8.57TLSGSDD1526 pKa = 3.61TIQVRR1531 pKa = 11.84LTDD1534 pKa = 3.76GTGVAGTPVSHH1545 pKa = 7.29TYY1547 pKa = 10.22TIDD1550 pKa = 3.33TTSPGSMSSKK1560 pKa = 10.39IPDD1563 pKa = 3.7LASSSDD1569 pKa = 3.28TSNGVSGGTSDD1580 pKa = 6.36DD1581 pKa = 3.42ITSNTTPVITYY1592 pKa = 10.16SLYY1595 pKa = 11.07NSGLTAGDD1603 pKa = 3.75YY1604 pKa = 11.15VDD1606 pKa = 5.57VYY1608 pKa = 11.09DD1609 pKa = 4.31TTTSTILHH1617 pKa = 5.71THH1619 pKa = 7.28TITSSEE1625 pKa = 4.04LTSYY1629 pKa = 10.99GGTVSATVSEE1639 pKa = 4.85LSAGSHH1645 pKa = 5.06SLVLRR1650 pKa = 11.84SRR1652 pKa = 11.84DD1653 pKa = 3.34AAGNTGTQSQALALTIDD1670 pKa = 3.94TSAISMAGKK1679 pKa = 8.12TVDD1682 pKa = 4.07LAASSDD1688 pKa = 3.84SGTSSTDD1695 pKa = 3.14NITNNSAPTVTVNVANTSGLVAGDD1719 pKa = 3.59KK1720 pKa = 10.83LQIIDD1725 pKa = 4.35SSNGDD1730 pKa = 3.64AVVGTYY1736 pKa = 9.34TLTSTDD1742 pKa = 4.05FSSSGADD1749 pKa = 3.13INITLSTLTDD1759 pKa = 3.51GTHH1762 pKa = 6.55ALALRR1767 pKa = 11.84VYY1769 pKa = 10.56DD1770 pKa = 3.6RR1771 pKa = 11.84AGNTGTASSATSITIDD1787 pKa = 3.81TIAPSLSSSTPSSSSTSNSVSTSSVTLVFSEE1818 pKa = 4.54ALADD1822 pKa = 3.79VNSRR1826 pKa = 11.84TGFKK1830 pKa = 9.55VTDD1833 pKa = 3.86GANDD1837 pKa = 3.17ARR1839 pKa = 11.84EE1840 pKa = 4.15ITGEE1844 pKa = 3.97ALSGNTLSYY1853 pKa = 11.2DD1854 pKa = 3.71SSTHH1858 pKa = 5.29TATLSLSGSLQYY1870 pKa = 11.47ASTYY1874 pKa = 8.08TVYY1877 pKa = 10.73VYY1879 pKa = 10.91NAALSDD1885 pKa = 3.56TAGNSVAIDD1894 pKa = 3.44TALLSFSTEE1903 pKa = 4.24SVPTPSAVTLAMTEE1917 pKa = 4.0STYY1920 pKa = 11.33ASHH1923 pKa = 7.16TGTDD1927 pKa = 3.42NDD1929 pKa = 5.27HH1930 pKa = 6.35ITNNATLCVSGITAAYY1946 pKa = 9.69LRR1948 pKa = 11.84YY1949 pKa = 10.06KK1950 pKa = 9.49LTSNAGWTTVQTSGSTYY1967 pKa = 10.41SIPLSDD1973 pKa = 3.97DD1974 pKa = 3.24TYY1976 pKa = 10.67AAGVIQVEE1984 pKa = 4.6QYY1986 pKa = 8.96DD1987 pKa = 3.95TAGYY1991 pKa = 9.77EE1992 pKa = 4.13STAATLSDD2000 pKa = 3.47SWTVDD2005 pKa = 2.83TSSPSGRR2012 pKa = 11.84ISTSTSGLPSFTGLGINTAISTITGSVSGEE2042 pKa = 3.87FNLDD2046 pKa = 3.5DD2047 pKa = 4.26EE2048 pKa = 5.28FIEE2051 pKa = 4.38YY2052 pKa = 7.59TTDD2055 pKa = 4.69SGSTWTKK2062 pKa = 10.18ASAMTAFIWTIADD2075 pKa = 3.39VKK2077 pKa = 10.34VAAGGEE2083 pKa = 3.94IGFRR2087 pKa = 11.84ASDD2090 pKa = 3.16KK2091 pKa = 11.0AGNIGYY2097 pKa = 10.03YY2098 pKa = 10.75GEE2100 pKa = 4.16TSNYY2104 pKa = 7.35TVYY2107 pKa = 11.01VGDD2110 pKa = 4.61DD2111 pKa = 3.06SAGNHH2116 pKa = 5.57TATGSKK2122 pKa = 10.34YY2123 pKa = 10.69LYY2125 pKa = 8.71TQAGNDD2131 pKa = 3.89TIDD2134 pKa = 3.0ITGASNAVHH2143 pKa = 6.81AGSGNDD2149 pKa = 3.85AININSASTTNHH2161 pKa = 5.5VYY2163 pKa = 11.14GDD2165 pKa = 4.07GGSDD2169 pKa = 3.55TINVNGSHH2177 pKa = 6.68NSQIEE2182 pKa = 4.07GGADD2186 pKa = 3.34ADD2188 pKa = 4.56TITVTGSYY2196 pKa = 10.64NYY2198 pKa = 10.38VYY2200 pKa = 10.98GNDD2203 pKa = 3.78GDD2205 pKa = 4.25DD2206 pKa = 3.6TLSISGTYY2214 pKa = 7.9NTVYY2218 pKa = 10.82GDD2220 pKa = 3.51NSEE2223 pKa = 4.33TGDD2226 pKa = 3.6TGADD2230 pKa = 3.63TITVSGSSNTVDD2242 pKa = 3.87AGPGANSITVTGADD2256 pKa = 3.31NTVTGGSDD2264 pKa = 3.1TDD2266 pKa = 4.18TISVSSTGNTVYY2278 pKa = 10.84GGDD2281 pKa = 3.65SADD2284 pKa = 4.02TITVATSGNTIYY2296 pKa = 11.23GEE2298 pKa = 4.15AGADD2302 pKa = 3.52QFTVAALGSNIYY2314 pKa = 10.67GGDD2317 pKa = 3.55GSDD2320 pKa = 3.33TFTVTASLAGTLSGLIDD2337 pKa = 4.31GGTDD2341 pKa = 3.18GTDD2344 pKa = 3.16VLNIGVSSQTLSLSTLTSNIAHH2366 pKa = 6.91IEE2368 pKa = 4.14KK2369 pKa = 10.33ISLGTSNTLTISGAAIQTMSDD2390 pKa = 2.89TDD2392 pKa = 4.0SIRR2395 pKa = 11.84FEE2397 pKa = 4.47GDD2399 pKa = 3.13STDD2402 pKa = 3.35TIYY2405 pKa = 11.33YY2406 pKa = 10.3DD2407 pKa = 3.33SSLWEE2412 pKa = 4.02SATWIQILGYY2422 pKa = 10.35NKK2424 pKa = 10.12YY2425 pKa = 8.42QLKK2428 pKa = 10.49SDD2430 pKa = 3.56TTITFIIDD2438 pKa = 3.45SDD2440 pKa = 3.31IHH2442 pKa = 8.31LYY2444 pKa = 11.12GLTSS2448 pKa = 3.21
MM1 pKa = 7.86SDD3 pKa = 3.39LTAPSLKK10 pKa = 10.52SVTFTNSTANASAVNTSADD29 pKa = 3.09IVLTFSEE36 pKa = 4.78AMTSGTGLITISDD49 pKa = 4.49GATQTYY55 pKa = 8.45MGKK58 pKa = 10.2DD59 pKa = 2.98GTLRR63 pKa = 11.84TRR65 pKa = 11.84LVGATDD71 pKa = 3.26TRR73 pKa = 11.84TVDD76 pKa = 3.62ISDD79 pKa = 3.8TSQVTVSGDD88 pKa = 3.24TVTINLASDD97 pKa = 3.96LKK99 pKa = 10.78AGKK102 pKa = 9.89SYY104 pKa = 10.56SVQMGKK110 pKa = 10.57GVLLDD115 pKa = 3.78EE116 pKa = 5.65AGNAYY121 pKa = 10.31AGLSDD126 pKa = 3.5TTKK129 pKa = 11.15LKK131 pKa = 9.6FTTSAATLPTAVVDD145 pKa = 3.86STFTITDD152 pKa = 3.69TGSSASDD159 pKa = 4.47FITSSAVQTIAGKK172 pKa = 10.95VNGALLAGEE181 pKa = 4.42AVMVSLDD188 pKa = 4.74NGGTWAAATVNAADD202 pKa = 4.14STWSIGGTVTGSSTIIARR220 pKa = 11.84VVNAEE225 pKa = 4.09GNASAAARR233 pKa = 11.84HH234 pKa = 6.32AYY236 pKa = 9.06TFDD239 pKa = 3.25TDD241 pKa = 3.38APTVGSIAINDD252 pKa = 3.68TDD254 pKa = 4.12LVAGEE259 pKa = 4.32TATVTVTFNEE269 pKa = 4.36AVTGLDD275 pKa = 3.13VSDD278 pKa = 3.95FTVEE282 pKa = 4.03GGALSSLATADD293 pKa = 4.31NGKK296 pKa = 7.63TWTATLTPTKK306 pKa = 10.29GVSTTTGTVTLNAGSVADD324 pKa = 4.0LAGNSGPANSSSKK337 pKa = 9.78PFNYY341 pKa = 8.93STVPDD346 pKa = 4.14APTATVTSISMTDD359 pKa = 3.3TGSSATDD366 pKa = 4.37FITNSTSQTIKK377 pKa = 9.49GTYY380 pKa = 9.73SGTLAADD387 pKa = 4.28EE388 pKa = 4.75YY389 pKa = 10.97IQVSLDD395 pKa = 3.7NGSTWTKK402 pKa = 9.82ATVSGSNWEE411 pKa = 4.12VAGATISASNTLKK424 pKa = 10.79ARR426 pKa = 11.84VFNDD430 pKa = 3.19YY431 pKa = 10.2TYY433 pKa = 11.08SNPMAQAYY441 pKa = 7.86TLDD444 pKa = 3.55QTAPTATVTLDD455 pKa = 3.09KK456 pKa = 11.42AHH458 pKa = 6.61LKK460 pKa = 10.47KK461 pKa = 11.16GEE463 pKa = 4.17TATITITFSEE473 pKa = 4.79AVTGLTSADD482 pKa = 3.03IKK484 pKa = 11.62SNDD487 pKa = 3.65TVGTPVSTDD496 pKa = 3.05GGVTWTATLTPTNNGNGSVTLYY518 pKa = 10.76ASSVTDD524 pKa = 3.5LAGNSGPAADD534 pKa = 3.82VAATYY539 pKa = 10.78SSDD542 pKa = 3.35LTRR545 pKa = 11.84PTATISLADD554 pKa = 3.5SALTAGEE561 pKa = 4.36TTLVTITFSEE571 pKa = 4.44AVTEE575 pKa = 4.19FTNADD580 pKa = 3.6LTVGNGTLSTVTSTDD595 pKa = 3.2GGTTWTATLTPTASTTATTNVITLDD620 pKa = 3.47NTAVSDD626 pKa = 3.84LAGNTGIGSTSSANYY641 pKa = 9.31SVSTVRR647 pKa = 11.84PTATINIADD656 pKa = 4.02TALTAGEE663 pKa = 4.41TTTVTFTFSSAVTGFDD679 pKa = 3.3ASDD682 pKa = 3.29VTVANGTISTPTTTDD697 pKa = 3.13NITWTATLTPSASTEE712 pKa = 3.85DD713 pKa = 3.56TTNVITLANTGFHH726 pKa = 6.5NSAGNSGTGTTTSGNYY742 pKa = 8.72TVDD745 pKa = 3.35TARR748 pKa = 11.84PTATISLADD757 pKa = 3.49TALTVGEE764 pKa = 4.51TTTVTVKK771 pKa = 10.46FSEE774 pKa = 4.31AVTGFSASSLSVANGTISSLTSSDD798 pKa = 3.47NITWTGTLTPTASQEE813 pKa = 3.91DD814 pKa = 4.19STNVITLNNSGVSDD828 pKa = 3.61AAGNTGSGTTTSANYY843 pKa = 9.02TVDD846 pKa = 3.31TLRR849 pKa = 11.84PTATISLADD858 pKa = 3.5SALTAGEE865 pKa = 4.35TTTVSITFSEE875 pKa = 5.01VVTGLTTADD884 pKa = 3.35LTVANGTLTSLQSSDD899 pKa = 3.04GMNWTATLTPSASTDD914 pKa = 3.14DD915 pKa = 3.77TTNVITLDD923 pKa = 3.35NTGITDD929 pKa = 3.63ASGNAGSGTTTSANYY944 pKa = 8.53TVATVRR950 pKa = 11.84PTATIAVSDD959 pKa = 4.02TALKK963 pKa = 9.91TGDD966 pKa = 3.6TATVTITFSEE976 pKa = 4.55AVTGLTAADD985 pKa = 3.35FTVANGTLSTPTTTDD1000 pKa = 3.09NITWTATLTPTSSIEE1015 pKa = 3.9DD1016 pKa = 3.71TTNVITLDD1024 pKa = 3.54NTGVTDD1030 pKa = 3.43SDD1032 pKa = 4.3GNTGSGTTSSSNYY1045 pKa = 9.21TVDD1048 pKa = 3.26TKK1050 pKa = 11.59APTATIALSQPEE1062 pKa = 4.87LLTGEE1067 pKa = 4.36TSTVTITFNEE1077 pKa = 4.26AVTGLTLADD1086 pKa = 4.31FKK1088 pKa = 11.72YY1089 pKa = 10.16SGTMTAPVSTDD1100 pKa = 3.26GGTTWVATLTPSTEE1114 pKa = 4.09SSSGEE1119 pKa = 3.72VRR1121 pKa = 11.84LNTSSVTDD1129 pKa = 3.37SAGNSGPSSEE1139 pKa = 5.13AYY1141 pKa = 10.35ASFTYY1146 pKa = 10.35NAAPTATIATASITDD1161 pKa = 3.64TGTSGDD1167 pKa = 4.69FITSSTSQEE1176 pKa = 3.56ISGTLSAALGRR1187 pKa = 11.84GQYY1190 pKa = 9.81VQVSLDD1196 pKa = 3.59NGEE1199 pKa = 4.36SWTTATTIGTGWSLSGQTISHH1220 pKa = 6.52SSTLKK1225 pKa = 9.98VRR1227 pKa = 11.84ATNGSSTSTTYY1238 pKa = 10.48SHH1240 pKa = 7.41SYY1242 pKa = 9.6TLDD1245 pKa = 3.04QAAPTAPTITLSKK1258 pKa = 10.57DD1259 pKa = 3.08ALYY1262 pKa = 10.33TNQTATVTVVFDD1274 pKa = 3.44EE1275 pKa = 4.4AVYY1278 pKa = 10.95GLTASDD1284 pKa = 3.62FTISGGTLGTFSTSDD1299 pKa = 3.33NLTWTATLTPTATGSLALAASSVYY1323 pKa = 9.49DD1324 pKa = 3.3TAGNVGPTSASSSKK1338 pKa = 10.94SFTFTADD1345 pKa = 3.1TTGPTATLTLNDD1357 pKa = 3.44NTLGKK1362 pKa = 10.69GGTATLTVTLSEE1374 pKa = 4.6AVTDD1378 pKa = 3.99TPTASDD1384 pKa = 3.7FTVSGGSLGTFTSTDD1399 pKa = 3.07SGLTWTATLTPTDD1412 pKa = 3.54SAYY1415 pKa = 10.5GAGTISLSANAISDD1429 pKa = 3.5QSGNTGPSSAASLPFYY1445 pKa = 10.9YY1446 pKa = 9.36DD1447 pKa = 3.27TQALTLSSSIAFSTDD1462 pKa = 2.79SGEE1465 pKa = 4.31SGSDD1469 pKa = 3.68LITNTGTQTISGTYY1483 pKa = 9.53SGGTIYY1489 pKa = 10.04STMGDD1494 pKa = 3.82KK1495 pKa = 10.7IEE1497 pKa = 4.01VSTDD1501 pKa = 3.07NGSTWSEE1508 pKa = 3.83ATVSGQAWSISKK1520 pKa = 8.57TLSGSDD1526 pKa = 3.61TIQVRR1531 pKa = 11.84LTDD1534 pKa = 3.76GTGVAGTPVSHH1545 pKa = 7.29TYY1547 pKa = 10.22TIDD1550 pKa = 3.33TTSPGSMSSKK1560 pKa = 10.39IPDD1563 pKa = 3.7LASSSDD1569 pKa = 3.28TSNGVSGGTSDD1580 pKa = 6.36DD1581 pKa = 3.42ITSNTTPVITYY1592 pKa = 10.16SLYY1595 pKa = 11.07NSGLTAGDD1603 pKa = 3.75YY1604 pKa = 11.15VDD1606 pKa = 5.57VYY1608 pKa = 11.09DD1609 pKa = 4.31TTTSTILHH1617 pKa = 5.71THH1619 pKa = 7.28TITSSEE1625 pKa = 4.04LTSYY1629 pKa = 10.99GGTVSATVSEE1639 pKa = 4.85LSAGSHH1645 pKa = 5.06SLVLRR1650 pKa = 11.84SRR1652 pKa = 11.84DD1653 pKa = 3.34AAGNTGTQSQALALTIDD1670 pKa = 3.94TSAISMAGKK1679 pKa = 8.12TVDD1682 pKa = 4.07LAASSDD1688 pKa = 3.84SGTSSTDD1695 pKa = 3.14NITNNSAPTVTVNVANTSGLVAGDD1719 pKa = 3.59KK1720 pKa = 10.83LQIIDD1725 pKa = 4.35SSNGDD1730 pKa = 3.64AVVGTYY1736 pKa = 9.34TLTSTDD1742 pKa = 4.05FSSSGADD1749 pKa = 3.13INITLSTLTDD1759 pKa = 3.51GTHH1762 pKa = 6.55ALALRR1767 pKa = 11.84VYY1769 pKa = 10.56DD1770 pKa = 3.6RR1771 pKa = 11.84AGNTGTASSATSITIDD1787 pKa = 3.81TIAPSLSSSTPSSSSTSNSVSTSSVTLVFSEE1818 pKa = 4.54ALADD1822 pKa = 3.79VNSRR1826 pKa = 11.84TGFKK1830 pKa = 9.55VTDD1833 pKa = 3.86GANDD1837 pKa = 3.17ARR1839 pKa = 11.84EE1840 pKa = 4.15ITGEE1844 pKa = 3.97ALSGNTLSYY1853 pKa = 11.2DD1854 pKa = 3.71SSTHH1858 pKa = 5.29TATLSLSGSLQYY1870 pKa = 11.47ASTYY1874 pKa = 8.08TVYY1877 pKa = 10.73VYY1879 pKa = 10.91NAALSDD1885 pKa = 3.56TAGNSVAIDD1894 pKa = 3.44TALLSFSTEE1903 pKa = 4.24SVPTPSAVTLAMTEE1917 pKa = 4.0STYY1920 pKa = 11.33ASHH1923 pKa = 7.16TGTDD1927 pKa = 3.42NDD1929 pKa = 5.27HH1930 pKa = 6.35ITNNATLCVSGITAAYY1946 pKa = 9.69LRR1948 pKa = 11.84YY1949 pKa = 10.06KK1950 pKa = 9.49LTSNAGWTTVQTSGSTYY1967 pKa = 10.41SIPLSDD1973 pKa = 3.97DD1974 pKa = 3.24TYY1976 pKa = 10.67AAGVIQVEE1984 pKa = 4.6QYY1986 pKa = 8.96DD1987 pKa = 3.95TAGYY1991 pKa = 9.77EE1992 pKa = 4.13STAATLSDD2000 pKa = 3.47SWTVDD2005 pKa = 2.83TSSPSGRR2012 pKa = 11.84ISTSTSGLPSFTGLGINTAISTITGSVSGEE2042 pKa = 3.87FNLDD2046 pKa = 3.5DD2047 pKa = 4.26EE2048 pKa = 5.28FIEE2051 pKa = 4.38YY2052 pKa = 7.59TTDD2055 pKa = 4.69SGSTWTKK2062 pKa = 10.18ASAMTAFIWTIADD2075 pKa = 3.39VKK2077 pKa = 10.34VAAGGEE2083 pKa = 3.94IGFRR2087 pKa = 11.84ASDD2090 pKa = 3.16KK2091 pKa = 11.0AGNIGYY2097 pKa = 10.03YY2098 pKa = 10.75GEE2100 pKa = 4.16TSNYY2104 pKa = 7.35TVYY2107 pKa = 11.01VGDD2110 pKa = 4.61DD2111 pKa = 3.06SAGNHH2116 pKa = 5.57TATGSKK2122 pKa = 10.34YY2123 pKa = 10.69LYY2125 pKa = 8.71TQAGNDD2131 pKa = 3.89TIDD2134 pKa = 3.0ITGASNAVHH2143 pKa = 6.81AGSGNDD2149 pKa = 3.85AININSASTTNHH2161 pKa = 5.5VYY2163 pKa = 11.14GDD2165 pKa = 4.07GGSDD2169 pKa = 3.55TINVNGSHH2177 pKa = 6.68NSQIEE2182 pKa = 4.07GGADD2186 pKa = 3.34ADD2188 pKa = 4.56TITVTGSYY2196 pKa = 10.64NYY2198 pKa = 10.38VYY2200 pKa = 10.98GNDD2203 pKa = 3.78GDD2205 pKa = 4.25DD2206 pKa = 3.6TLSISGTYY2214 pKa = 7.9NTVYY2218 pKa = 10.82GDD2220 pKa = 3.51NSEE2223 pKa = 4.33TGDD2226 pKa = 3.6TGADD2230 pKa = 3.63TITVSGSSNTVDD2242 pKa = 3.87AGPGANSITVTGADD2256 pKa = 3.31NTVTGGSDD2264 pKa = 3.1TDD2266 pKa = 4.18TISVSSTGNTVYY2278 pKa = 10.84GGDD2281 pKa = 3.65SADD2284 pKa = 4.02TITVATSGNTIYY2296 pKa = 11.23GEE2298 pKa = 4.15AGADD2302 pKa = 3.52QFTVAALGSNIYY2314 pKa = 10.67GGDD2317 pKa = 3.55GSDD2320 pKa = 3.33TFTVTASLAGTLSGLIDD2337 pKa = 4.31GGTDD2341 pKa = 3.18GTDD2344 pKa = 3.16VLNIGVSSQTLSLSTLTSNIAHH2366 pKa = 6.91IEE2368 pKa = 4.14KK2369 pKa = 10.33ISLGTSNTLTISGAAIQTMSDD2390 pKa = 2.89TDD2392 pKa = 4.0SIRR2395 pKa = 11.84FEE2397 pKa = 4.47GDD2399 pKa = 3.13STDD2402 pKa = 3.35TIYY2405 pKa = 11.33YY2406 pKa = 10.3DD2407 pKa = 3.33SSLWEE2412 pKa = 4.02SATWIQILGYY2422 pKa = 10.35NKK2424 pKa = 10.12YY2425 pKa = 8.42QLKK2428 pKa = 10.49SDD2430 pKa = 3.56TTITFIIDD2438 pKa = 3.45SDD2440 pKa = 3.31IHH2442 pKa = 8.31LYY2444 pKa = 11.12GLTSS2448 pKa = 3.21
Molecular weight: 246.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L6Q7R1|A0A6L6Q7R1_9BURK Uncharacterized protein OS=Duganella ginsengisoli OX=1462440 GN=GM668_27725 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84QVLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LAVVV45 pKa = 3.61
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84QVLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LAVVV45 pKa = 3.61
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2124225 |
37 |
12803 |
358.9 |
38.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.019 ± 0.053 | 0.883 ± 0.011 |
5.416 ± 0.023 | 4.965 ± 0.034 |
3.404 ± 0.02 | 8.152 ± 0.041 |
2.235 ± 0.019 | 4.56 ± 0.025 |
3.676 ± 0.031 | 10.305 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.638 ± 0.018 | 3.149 ± 0.025 |
4.916 ± 0.03 | 4.261 ± 0.028 |
6.155 ± 0.042 | 5.571 ± 0.036 |
5.439 ± 0.05 | 7.301 ± 0.034 |
1.325 ± 0.014 | 2.629 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
