
Sulfolobus islandicus filamentous virus (isolate Iceland/Hveragerdi) (SIFV)
Taxonomy: Viruses; Adnaviria; Zilligvirae; Taleaviricota; Tokiviricetes; Ligamenvirales; Lipothrixviridae; Betalipothrixvirus; Sulfolobus islandicus filamentous virus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
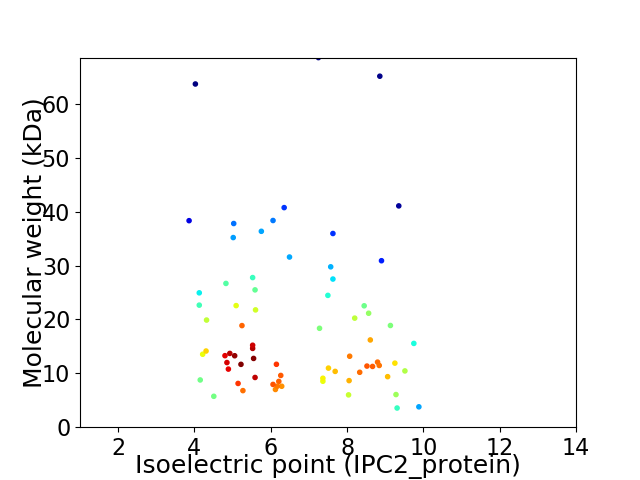
Virtual 2D-PAGE plot for 73 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
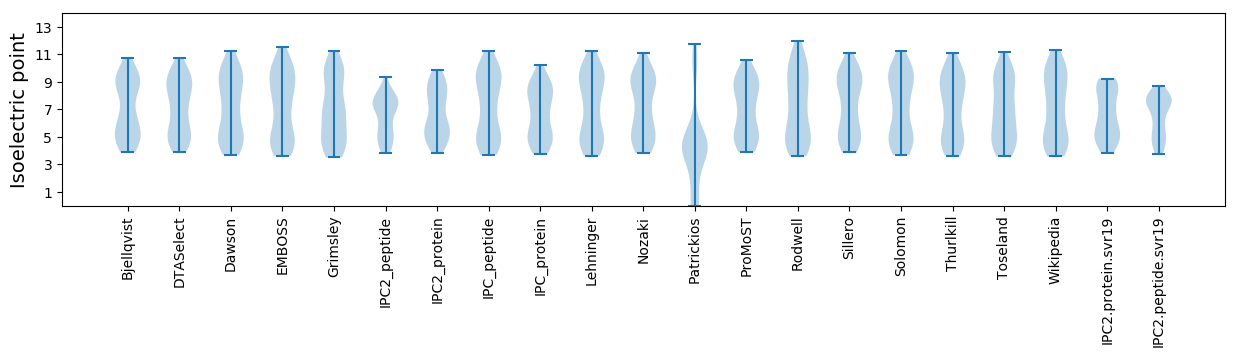
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
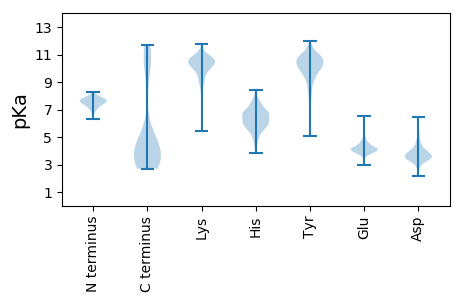
>sp|Q914H7|Y055_SIFVH Uncharacterized protein 55 OS=Sulfolobus islandicus filamentous virus (isolate Iceland/Hveragerdi) OX=654908 GN=SIFV0055 PE=4 SV=1
MM1 pKa = 7.77SFLLNLGQAFFSGLSSIATGVTSDD25 pKa = 3.51VTSVANGINYY35 pKa = 9.13AFNVLANFLQKK46 pKa = 10.62LPSEE50 pKa = 4.11IATFFQSIPGALIGFAHH67 pKa = 6.79TFGTYY72 pKa = 8.77IWDD75 pKa = 3.55GMQRR79 pKa = 11.84IASGFTAIMAPIEE92 pKa = 4.5RR93 pKa = 11.84GLEE96 pKa = 4.05TLGTTVINALTVVWNDD112 pKa = 2.43IKK114 pKa = 11.33AFASAIYY121 pKa = 10.3NSIVSMINNIVSFVQPFINDD141 pKa = 3.83FKK143 pKa = 11.04NAISFAYY150 pKa = 10.29NFLLNVINDD159 pKa = 3.49VYY161 pKa = 11.42SAFTIISNFFLDD173 pKa = 5.26LPTFFQNASNYY184 pKa = 10.02LNSLFTDD191 pKa = 4.43PGNQPNLLTMVPNLVASEE209 pKa = 4.07VSRR212 pKa = 11.84IAGAFPDD219 pKa = 4.22VIAYY223 pKa = 6.7NTFMEE228 pKa = 4.74VLPKK232 pKa = 9.45MVSGIASSPIFGNTVKK248 pKa = 10.84GMFAKK253 pKa = 10.37ALLMAGSPILSALLSEE269 pKa = 4.33LTKK272 pKa = 11.29ALMQSLFTSTQTTQTTQRR290 pKa = 11.84PSQPQIQPPTAPSTQLQQRR309 pKa = 11.84STQQTSLSDD318 pKa = 3.65LQNLQTQQLTPTQIEE333 pKa = 4.49VQLEE337 pKa = 4.24RR338 pKa = 11.84PNVTGVFTQDD348 pKa = 3.36VIGMGTASGGTAKK361 pKa = 10.79LVTGYY366 pKa = 10.62INFQNSIKK374 pKa = 10.46QFEE377 pKa = 4.59DD378 pKa = 3.39VFNITASFFMEE389 pKa = 5.74LIQSLQTEE397 pKa = 5.28FSQDD401 pKa = 2.89MAVNVSLTLLEE412 pKa = 4.75NIYY415 pKa = 10.34QVSQSIEE422 pKa = 3.96IIPSVDD428 pKa = 2.73ATTIILPPGISLCNPSQAPSPSEE451 pKa = 4.02STSTSTNSIDD461 pKa = 3.18VTTTVDD467 pKa = 3.42VTEE470 pKa = 4.49EE471 pKa = 4.14LCIPPYY477 pKa = 10.83DD478 pKa = 4.18LLADD482 pKa = 4.18GLTTQFAVIALLHH495 pKa = 6.18SNISDD500 pKa = 3.32VMLAITQLIYY510 pKa = 10.82GITFSNLISDD520 pKa = 4.29ILSATTQLMYY530 pKa = 10.6SITSNISTSDD540 pKa = 3.43TVSVSASFTPSPPPSEE556 pKa = 4.12TYY558 pKa = 9.46TYY560 pKa = 10.55YY561 pKa = 11.02VPIDD565 pKa = 3.28IAVTYY570 pKa = 7.37TIQVASSTTYY580 pKa = 10.68SGALSYY586 pKa = 10.07TVSTSS591 pKa = 2.73
MM1 pKa = 7.77SFLLNLGQAFFSGLSSIATGVTSDD25 pKa = 3.51VTSVANGINYY35 pKa = 9.13AFNVLANFLQKK46 pKa = 10.62LPSEE50 pKa = 4.11IATFFQSIPGALIGFAHH67 pKa = 6.79TFGTYY72 pKa = 8.77IWDD75 pKa = 3.55GMQRR79 pKa = 11.84IASGFTAIMAPIEE92 pKa = 4.5RR93 pKa = 11.84GLEE96 pKa = 4.05TLGTTVINALTVVWNDD112 pKa = 2.43IKK114 pKa = 11.33AFASAIYY121 pKa = 10.3NSIVSMINNIVSFVQPFINDD141 pKa = 3.83FKK143 pKa = 11.04NAISFAYY150 pKa = 10.29NFLLNVINDD159 pKa = 3.49VYY161 pKa = 11.42SAFTIISNFFLDD173 pKa = 5.26LPTFFQNASNYY184 pKa = 10.02LNSLFTDD191 pKa = 4.43PGNQPNLLTMVPNLVASEE209 pKa = 4.07VSRR212 pKa = 11.84IAGAFPDD219 pKa = 4.22VIAYY223 pKa = 6.7NTFMEE228 pKa = 4.74VLPKK232 pKa = 9.45MVSGIASSPIFGNTVKK248 pKa = 10.84GMFAKK253 pKa = 10.37ALLMAGSPILSALLSEE269 pKa = 4.33LTKK272 pKa = 11.29ALMQSLFTSTQTTQTTQRR290 pKa = 11.84PSQPQIQPPTAPSTQLQQRR309 pKa = 11.84STQQTSLSDD318 pKa = 3.65LQNLQTQQLTPTQIEE333 pKa = 4.49VQLEE337 pKa = 4.24RR338 pKa = 11.84PNVTGVFTQDD348 pKa = 3.36VIGMGTASGGTAKK361 pKa = 10.79LVTGYY366 pKa = 10.62INFQNSIKK374 pKa = 10.46QFEE377 pKa = 4.59DD378 pKa = 3.39VFNITASFFMEE389 pKa = 5.74LIQSLQTEE397 pKa = 5.28FSQDD401 pKa = 2.89MAVNVSLTLLEE412 pKa = 4.75NIYY415 pKa = 10.34QVSQSIEE422 pKa = 3.96IIPSVDD428 pKa = 2.73ATTIILPPGISLCNPSQAPSPSEE451 pKa = 4.02STSTSTNSIDD461 pKa = 3.18VTTTVDD467 pKa = 3.42VTEE470 pKa = 4.49EE471 pKa = 4.14LCIPPYY477 pKa = 10.83DD478 pKa = 4.18LLADD482 pKa = 4.18GLTTQFAVIALLHH495 pKa = 6.18SNISDD500 pKa = 3.32VMLAITQLIYY510 pKa = 10.82GITFSNLISDD520 pKa = 4.29ILSATTQLMYY530 pKa = 10.6SITSNISTSDD540 pKa = 3.43TVSVSASFTPSPPPSEE556 pKa = 4.12TYY558 pKa = 9.46TYY560 pKa = 10.55YY561 pKa = 11.02VPIDD565 pKa = 3.28IAVTYY570 pKa = 7.37TIQVASSTTYY580 pKa = 10.68SGALSYY586 pKa = 10.07TVSTSS591 pKa = 2.73
Molecular weight: 63.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q914I4|GT048_SIFVH Putative glycosyltransferase 48 OS=Sulfolobus islandicus filamentous virus (isolate Iceland/Hveragerdi) OX=654908 GN=SIFV0048 PE=3 SV=1
MM1 pKa = 7.04VRR3 pKa = 11.84ALLFHH8 pKa = 6.84VITYY12 pKa = 7.58TKK14 pKa = 10.39FIVPVVKK21 pKa = 10.35LLIMSAIAGVIAGAFGGGIGGVGDD45 pKa = 5.17AIGTIIGDD53 pKa = 3.99LEE55 pKa = 3.76RR56 pKa = 11.84AIARR60 pKa = 11.84FGGSIVNAFKK70 pKa = 10.4TVIDD74 pKa = 4.82KK75 pKa = 10.83ILTLAVRR82 pKa = 11.84IGRR85 pKa = 11.84IIEE88 pKa = 4.01KK89 pKa = 8.88YY90 pKa = 9.9FRR92 pKa = 11.84IAVHH96 pKa = 6.0YY97 pKa = 9.95IVLFLRR103 pKa = 11.84LAYY106 pKa = 10.06RR107 pKa = 11.84YY108 pKa = 7.88MYY110 pKa = 10.8SFYY113 pKa = 10.92TEE115 pKa = 3.97FQKK118 pKa = 11.18DD119 pKa = 3.44PWRR122 pKa = 11.84SLQFVGSMAILLNNSLFPP140 pKa = 4.81
MM1 pKa = 7.04VRR3 pKa = 11.84ALLFHH8 pKa = 6.84VITYY12 pKa = 7.58TKK14 pKa = 10.39FIVPVVKK21 pKa = 10.35LLIMSAIAGVIAGAFGGGIGGVGDD45 pKa = 5.17AIGTIIGDD53 pKa = 3.99LEE55 pKa = 3.76RR56 pKa = 11.84AIARR60 pKa = 11.84FGGSIVNAFKK70 pKa = 10.4TVIDD74 pKa = 4.82KK75 pKa = 10.83ILTLAVRR82 pKa = 11.84IGRR85 pKa = 11.84IIEE88 pKa = 4.01KK89 pKa = 8.88YY90 pKa = 9.9FRR92 pKa = 11.84IAVHH96 pKa = 6.0YY97 pKa = 9.95IVLFLRR103 pKa = 11.84LAYY106 pKa = 10.06RR107 pKa = 11.84YY108 pKa = 7.88MYY110 pKa = 10.8SFYY113 pKa = 10.92TEE115 pKa = 3.97FQKK118 pKa = 11.18DD119 pKa = 3.44PWRR122 pKa = 11.84SLQFVGSMAILLNNSLFPP140 pKa = 4.81
Molecular weight: 15.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12208 |
31 |
601 |
167.2 |
19.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.595 ± 0.299 | 1.204 ± 0.168 |
4.481 ± 0.242 | 6.873 ± 0.471 |
4.546 ± 0.216 | 4.718 ± 0.193 |
1.466 ± 0.144 | 9.469 ± 0.349 |
7.831 ± 0.597 | 9.092 ± 0.23 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.531 ± 0.114 | 6.176 ± 0.294 |
3.408 ± 0.222 | 3.268 ± 0.304 |
3.53 ± 0.288 | 6.758 ± 0.49 |
6.184 ± 0.503 | 7.298 ± 0.341 |
0.836 ± 0.106 | 5.734 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
