
Otarine picobirnavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Picobirnaviridae; Picobirnavirus; unclassified Picobirnavirus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
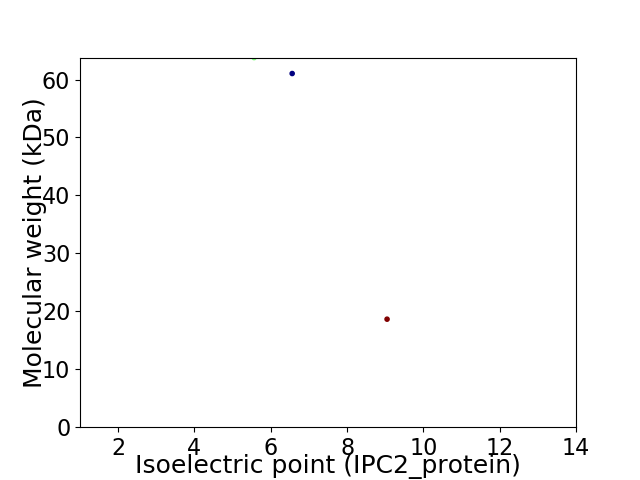
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I2DA78|I2DA78_9VIRU Putative RNA-dependent RNA polymerase OS=Otarine picobirnavirus OX=1187973 PE=4 SV=1
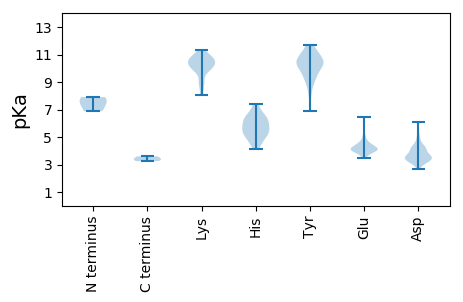
MM1 pKa = 7.54SKK3 pKa = 9.84MSTKK7 pKa = 10.5DD8 pKa = 3.29RR9 pKa = 11.84NNDD12 pKa = 3.35SLNGGTNNNARR23 pKa = 11.84QKK25 pKa = 10.97SNNKK29 pKa = 9.14RR30 pKa = 11.84RR31 pKa = 11.84GSKK34 pKa = 10.16NPNRR38 pKa = 11.84PSTRR42 pKa = 11.84NHH44 pKa = 5.86VGEE47 pKa = 4.43DD48 pKa = 3.23QHH50 pKa = 6.12YY51 pKa = 8.2TLANAPDD58 pKa = 4.29AGNSIDD64 pKa = 4.29WYY66 pKa = 10.21NKK68 pKa = 10.39SPVLLKK74 pKa = 10.72DD75 pKa = 3.23AANITMAHH83 pKa = 6.01PVGLGMSLVMPQGTSGKK100 pKa = 9.89VGTYY104 pKa = 9.93SDD106 pKa = 4.17PGIMALNFIPTIASGSSGTHH126 pKa = 5.8ASDD129 pKa = 3.61INSATNIAARR139 pKa = 11.84NIYY142 pKa = 10.55SFVRR146 pKa = 11.84HH147 pKa = 5.79TNSGHH152 pKa = 6.46ANYY155 pKa = 9.02DD156 pKa = 3.31APDD159 pKa = 3.43LMKK162 pKa = 10.89YY163 pKa = 10.1LIAMDD168 pKa = 4.91SIYY171 pKa = 10.3MVYY174 pKa = 10.31AHH176 pKa = 7.41CARR179 pKa = 11.84IYY181 pKa = 10.83GVLRR185 pKa = 11.84LYY187 pKa = 10.74DD188 pKa = 3.5GRR190 pKa = 11.84NRR192 pKa = 11.84YY193 pKa = 9.27YY194 pKa = 9.99PDD196 pKa = 3.39QILRR200 pKa = 11.84ALGVANVTPQMGINEE215 pKa = 4.09FRR217 pKa = 11.84SALNRR222 pKa = 11.84IAYY225 pKa = 9.04KK226 pKa = 10.05IGVMNVPNNLSYY238 pKa = 9.57MQRR241 pKa = 11.84HH242 pKa = 5.44MWLCSNLYY250 pKa = 10.47ADD252 pKa = 4.25EE253 pKa = 5.23PGNTFQTYY261 pKa = 9.2CFRR264 pKa = 11.84PGGAYY269 pKa = 9.53RR270 pKa = 11.84YY271 pKa = 9.76RR272 pKa = 11.84FSTPKK277 pKa = 10.7DD278 pKa = 3.39PTGSLEE284 pKa = 4.23MKK286 pKa = 10.12VLPTMSLQSWLNTLEE301 pKa = 4.18EE302 pKa = 4.11LMAPVLSSEE311 pKa = 4.73DD312 pKa = 3.59MAIMSGDD319 pKa = 3.58LLKK322 pKa = 10.78AYY324 pKa = 10.19GEE326 pKa = 4.12SSMFKK331 pKa = 9.7LTTVPEE337 pKa = 4.74DD338 pKa = 3.68YY339 pKa = 10.58QVLPTYY345 pKa = 10.56NEE347 pKa = 3.98EE348 pKa = 3.96VLSQIEE354 pKa = 4.44NATIWYY360 pKa = 9.4GIDD363 pKa = 3.44NSVEE367 pKa = 4.12SADD370 pKa = 4.02KK371 pKa = 10.87DD372 pKa = 3.42HH373 pKa = 6.99HH374 pKa = 6.37KK375 pKa = 10.5WDD377 pKa = 3.22IRR379 pKa = 11.84EE380 pKa = 4.01NTTQDD385 pKa = 3.63DD386 pKa = 4.33ANLGALYY393 pKa = 10.18QNPTLLSPYY402 pKa = 9.44GCQFDD407 pKa = 4.71SVINLHH413 pKa = 5.38TEE415 pKa = 3.79EE416 pKa = 4.39VNPEE420 pKa = 3.63NVMIATRR427 pKa = 11.84LMTTAVSAKK436 pKa = 8.28WMKK439 pKa = 10.0TKK441 pKa = 10.57EE442 pKa = 4.04KK443 pKa = 9.8LYY445 pKa = 10.77TMTAMDD451 pKa = 4.43YY452 pKa = 10.46GSEE455 pKa = 4.11IIVAAFMYY463 pKa = 9.84TGLGSANIDD472 pKa = 3.89DD473 pKa = 4.69LNCYY477 pKa = 10.18VDD479 pKa = 4.09IFSDD483 pKa = 3.67TTPAQAVSEE492 pKa = 4.47TAVSMIAAGYY502 pKa = 8.2LQTFNRR508 pKa = 11.84HH509 pKa = 5.14PLVFVGTRR517 pKa = 11.84TFKK520 pKa = 11.12DD521 pKa = 3.36PASTTTPSEE530 pKa = 3.83PTIYY534 pKa = 10.26TPAFMLGVSNMSTVDD549 pKa = 3.19HH550 pKa = 6.44RR551 pKa = 11.84TLEE554 pKa = 4.26SMHH557 pKa = 5.97NVAMLSMFDD566 pKa = 3.49VSSNNLNRR574 pKa = 11.84GG575 pKa = 3.25
MM1 pKa = 7.54SKK3 pKa = 9.84MSTKK7 pKa = 10.5DD8 pKa = 3.29RR9 pKa = 11.84NNDD12 pKa = 3.35SLNGGTNNNARR23 pKa = 11.84QKK25 pKa = 10.97SNNKK29 pKa = 9.14RR30 pKa = 11.84RR31 pKa = 11.84GSKK34 pKa = 10.16NPNRR38 pKa = 11.84PSTRR42 pKa = 11.84NHH44 pKa = 5.86VGEE47 pKa = 4.43DD48 pKa = 3.23QHH50 pKa = 6.12YY51 pKa = 8.2TLANAPDD58 pKa = 4.29AGNSIDD64 pKa = 4.29WYY66 pKa = 10.21NKK68 pKa = 10.39SPVLLKK74 pKa = 10.72DD75 pKa = 3.23AANITMAHH83 pKa = 6.01PVGLGMSLVMPQGTSGKK100 pKa = 9.89VGTYY104 pKa = 9.93SDD106 pKa = 4.17PGIMALNFIPTIASGSSGTHH126 pKa = 5.8ASDD129 pKa = 3.61INSATNIAARR139 pKa = 11.84NIYY142 pKa = 10.55SFVRR146 pKa = 11.84HH147 pKa = 5.79TNSGHH152 pKa = 6.46ANYY155 pKa = 9.02DD156 pKa = 3.31APDD159 pKa = 3.43LMKK162 pKa = 10.89YY163 pKa = 10.1LIAMDD168 pKa = 4.91SIYY171 pKa = 10.3MVYY174 pKa = 10.31AHH176 pKa = 7.41CARR179 pKa = 11.84IYY181 pKa = 10.83GVLRR185 pKa = 11.84LYY187 pKa = 10.74DD188 pKa = 3.5GRR190 pKa = 11.84NRR192 pKa = 11.84YY193 pKa = 9.27YY194 pKa = 9.99PDD196 pKa = 3.39QILRR200 pKa = 11.84ALGVANVTPQMGINEE215 pKa = 4.09FRR217 pKa = 11.84SALNRR222 pKa = 11.84IAYY225 pKa = 9.04KK226 pKa = 10.05IGVMNVPNNLSYY238 pKa = 9.57MQRR241 pKa = 11.84HH242 pKa = 5.44MWLCSNLYY250 pKa = 10.47ADD252 pKa = 4.25EE253 pKa = 5.23PGNTFQTYY261 pKa = 9.2CFRR264 pKa = 11.84PGGAYY269 pKa = 9.53RR270 pKa = 11.84YY271 pKa = 9.76RR272 pKa = 11.84FSTPKK277 pKa = 10.7DD278 pKa = 3.39PTGSLEE284 pKa = 4.23MKK286 pKa = 10.12VLPTMSLQSWLNTLEE301 pKa = 4.18EE302 pKa = 4.11LMAPVLSSEE311 pKa = 4.73DD312 pKa = 3.59MAIMSGDD319 pKa = 3.58LLKK322 pKa = 10.78AYY324 pKa = 10.19GEE326 pKa = 4.12SSMFKK331 pKa = 9.7LTTVPEE337 pKa = 4.74DD338 pKa = 3.68YY339 pKa = 10.58QVLPTYY345 pKa = 10.56NEE347 pKa = 3.98EE348 pKa = 3.96VLSQIEE354 pKa = 4.44NATIWYY360 pKa = 9.4GIDD363 pKa = 3.44NSVEE367 pKa = 4.12SADD370 pKa = 4.02KK371 pKa = 10.87DD372 pKa = 3.42HH373 pKa = 6.99HH374 pKa = 6.37KK375 pKa = 10.5WDD377 pKa = 3.22IRR379 pKa = 11.84EE380 pKa = 4.01NTTQDD385 pKa = 3.63DD386 pKa = 4.33ANLGALYY393 pKa = 10.18QNPTLLSPYY402 pKa = 9.44GCQFDD407 pKa = 4.71SVINLHH413 pKa = 5.38TEE415 pKa = 3.79EE416 pKa = 4.39VNPEE420 pKa = 3.63NVMIATRR427 pKa = 11.84LMTTAVSAKK436 pKa = 8.28WMKK439 pKa = 10.0TKK441 pKa = 10.57EE442 pKa = 4.04KK443 pKa = 9.8LYY445 pKa = 10.77TMTAMDD451 pKa = 4.43YY452 pKa = 10.46GSEE455 pKa = 4.11IIVAAFMYY463 pKa = 9.84TGLGSANIDD472 pKa = 3.89DD473 pKa = 4.69LNCYY477 pKa = 10.18VDD479 pKa = 4.09IFSDD483 pKa = 3.67TTPAQAVSEE492 pKa = 4.47TAVSMIAAGYY502 pKa = 8.2LQTFNRR508 pKa = 11.84HH509 pKa = 5.14PLVFVGTRR517 pKa = 11.84TFKK520 pKa = 11.12DD521 pKa = 3.36PASTTTPSEE530 pKa = 3.83PTIYY534 pKa = 10.26TPAFMLGVSNMSTVDD549 pKa = 3.19HH550 pKa = 6.44RR551 pKa = 11.84TLEE554 pKa = 4.26SMHH557 pKa = 5.97NVAMLSMFDD566 pKa = 3.49VSSNNLNRR574 pKa = 11.84GG575 pKa = 3.25
Molecular weight: 63.78 kDa
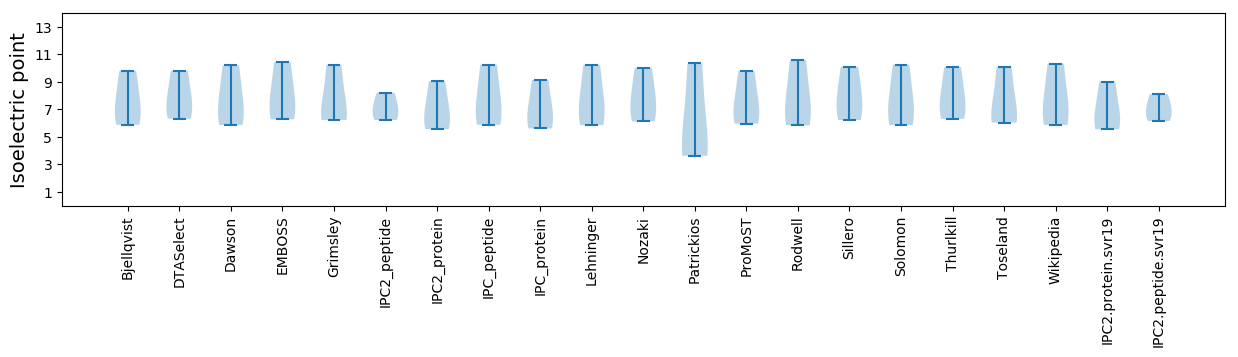
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I2DA77|I2DA77_9VIRU Capsid protein OS=Otarine picobirnavirus OX=1187973 GN=ORF2 PE=4 SV=1
MM1 pKa = 6.88TTNQINFFRR10 pKa = 11.84TQNEE14 pKa = 3.57KK15 pKa = 10.6LAAEE19 pKa = 4.85RR20 pKa = 11.84SAQQHH25 pKa = 4.52EE26 pKa = 4.54RR27 pKa = 11.84EE28 pKa = 4.25LNEE31 pKa = 3.48IRR33 pKa = 11.84RR34 pKa = 11.84SNLAKK39 pKa = 8.67EE40 pKa = 4.13QEE42 pKa = 4.08MRR44 pKa = 11.84RR45 pKa = 11.84SNMAKK50 pKa = 10.16EE51 pKa = 4.02NLGFQQFNEE60 pKa = 4.28SLRR63 pKa = 11.84HH64 pKa = 4.99NKK66 pKa = 8.16ATEE69 pKa = 4.15SQASSQLRR77 pKa = 11.84EE78 pKa = 3.97IEE80 pKa = 4.01RR81 pKa = 11.84HH82 pKa = 4.69NRR84 pKa = 11.84ASVGLGYY91 pKa = 11.05SNLSEE96 pKa = 3.96QHH98 pKa = 5.23RR99 pKa = 11.84HH100 pKa = 4.47NVASNSLGYY109 pKa = 10.99ANQFEE114 pKa = 4.7QNRR117 pKa = 11.84HH118 pKa = 5.38DD119 pKa = 3.69VAQEE123 pKa = 4.07LEE125 pKa = 4.42SQRR128 pKa = 11.84HH129 pKa = 4.15NTTTEE134 pKa = 3.66QTSNFGTIGNVLLGVGGLFTKK155 pKa = 10.46SNKK158 pKa = 9.05KK159 pKa = 10.05LKK161 pKa = 10.41RR162 pKa = 3.6
MM1 pKa = 6.88TTNQINFFRR10 pKa = 11.84TQNEE14 pKa = 3.57KK15 pKa = 10.6LAAEE19 pKa = 4.85RR20 pKa = 11.84SAQQHH25 pKa = 4.52EE26 pKa = 4.54RR27 pKa = 11.84EE28 pKa = 4.25LNEE31 pKa = 3.48IRR33 pKa = 11.84RR34 pKa = 11.84SNLAKK39 pKa = 8.67EE40 pKa = 4.13QEE42 pKa = 4.08MRR44 pKa = 11.84RR45 pKa = 11.84SNMAKK50 pKa = 10.16EE51 pKa = 4.02NLGFQQFNEE60 pKa = 4.28SLRR63 pKa = 11.84HH64 pKa = 4.99NKK66 pKa = 8.16ATEE69 pKa = 4.15SQASSQLRR77 pKa = 11.84EE78 pKa = 3.97IEE80 pKa = 4.01RR81 pKa = 11.84HH82 pKa = 4.69NRR84 pKa = 11.84ASVGLGYY91 pKa = 11.05SNLSEE96 pKa = 3.96QHH98 pKa = 5.23RR99 pKa = 11.84HH100 pKa = 4.47NVASNSLGYY109 pKa = 10.99ANQFEE114 pKa = 4.7QNRR117 pKa = 11.84HH118 pKa = 5.38DD119 pKa = 3.69VAQEE123 pKa = 4.07LEE125 pKa = 4.42SQRR128 pKa = 11.84HH129 pKa = 4.15NTTTEE134 pKa = 3.66QTSNFGTIGNVLLGVGGLFTKK155 pKa = 10.46SNKK158 pKa = 9.05KK159 pKa = 10.05LKK161 pKa = 10.41RR162 pKa = 3.6
Molecular weight: 18.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1269 |
162 |
575 |
423.0 |
47.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.698 ± 0.836 | 1.182 ± 0.407 |
6.225 ± 1.567 | 4.886 ± 1.437 |
3.31 ± 0.455 | 6.147 ± 0.168 |
2.522 ± 0.456 | 4.413 ± 0.513 |
5.122 ± 0.95 | 7.723 ± 0.231 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.255 ± 0.823 | 6.698 ± 1.879 |
4.571 ± 1.146 | 4.728 ± 1.542 |
5.674 ± 1.081 | 7.565 ± 0.937 |
6.619 ± 0.856 | 5.91 ± 0.819 |
1.34 ± 0.437 | 4.413 ± 0.85 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
