
Bacteroides coprophilus CAG:333
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
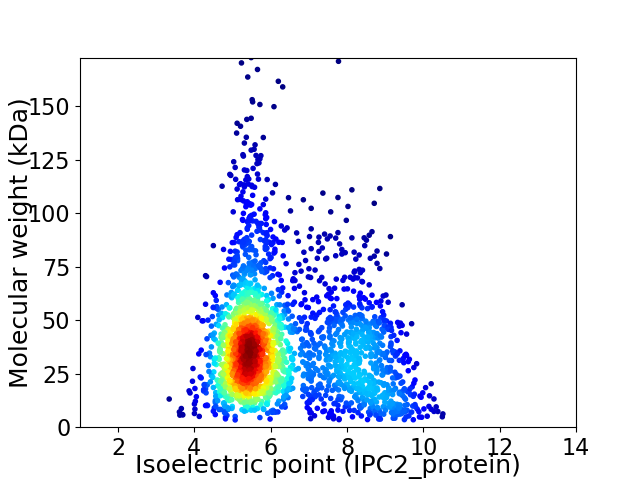
Virtual 2D-PAGE plot for 2352 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6TJR5|R6TJR5_9BACE DUF4105 domain-containing protein OS=Bacteroides coprophilus CAG:333 OX=1263041 GN=BN612_00957 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.22IKK4 pKa = 10.61GYY6 pKa = 10.29ILGISVAMASLLVSSCDD23 pKa = 3.35TDD25 pKa = 3.45NLTDD29 pKa = 4.0VYY31 pKa = 10.95QSSTIGATFITASQSVSFPSSGYY54 pKa = 9.71EE55 pKa = 3.92GFDD58 pKa = 3.45VEE60 pKa = 4.69VVRR63 pKa = 11.84AQTEE67 pKa = 3.95APATIKK73 pKa = 9.41VTSAVLLDD81 pKa = 3.91ADD83 pKa = 4.76GNALPLPSTIQVPSEE98 pKa = 4.12VNFEE102 pKa = 4.23AGEE105 pKa = 4.37GRR107 pKa = 11.84SSLHH111 pKa = 5.91VSVGDD116 pKa = 3.43ITPGEE121 pKa = 4.38TYY123 pKa = 10.62QLAITLDD130 pKa = 3.67EE131 pKa = 4.28TVAPVDD137 pKa = 3.24ATMTKK142 pKa = 9.78IISIFRR148 pKa = 11.84DD149 pKa = 3.37YY150 pKa = 10.92TYY152 pKa = 10.91TSLGTGTLNSTFFEE166 pKa = 4.37ATGTAEE172 pKa = 3.95IQKK175 pKa = 10.51ADD177 pKa = 3.25QTTWYY182 pKa = 9.71KK183 pKa = 11.03AIGLYY188 pKa = 10.41EE189 pKa = 4.21EE190 pKa = 5.69GYY192 pKa = 10.95DD193 pKa = 4.52IIFKK197 pKa = 10.13VAQDD201 pKa = 3.77GKK203 pKa = 9.07TVTVDD208 pKa = 3.27QQAAASDD215 pKa = 3.24ISGYY219 pKa = 8.37GTLYY223 pKa = 10.51VAGSGTLEE231 pKa = 3.97NGVITVEE238 pKa = 4.66LEE240 pKa = 4.08FTVSAGSFGSCTEE253 pKa = 3.87EE254 pKa = 5.47LILPAANN261 pKa = 3.27
MM1 pKa = 7.61KK2 pKa = 10.22IKK4 pKa = 10.61GYY6 pKa = 10.29ILGISVAMASLLVSSCDD23 pKa = 3.35TDD25 pKa = 3.45NLTDD29 pKa = 4.0VYY31 pKa = 10.95QSSTIGATFITASQSVSFPSSGYY54 pKa = 9.71EE55 pKa = 3.92GFDD58 pKa = 3.45VEE60 pKa = 4.69VVRR63 pKa = 11.84AQTEE67 pKa = 3.95APATIKK73 pKa = 9.41VTSAVLLDD81 pKa = 3.91ADD83 pKa = 4.76GNALPLPSTIQVPSEE98 pKa = 4.12VNFEE102 pKa = 4.23AGEE105 pKa = 4.37GRR107 pKa = 11.84SSLHH111 pKa = 5.91VSVGDD116 pKa = 3.43ITPGEE121 pKa = 4.38TYY123 pKa = 10.62QLAITLDD130 pKa = 3.67EE131 pKa = 4.28TVAPVDD137 pKa = 3.24ATMTKK142 pKa = 9.78IISIFRR148 pKa = 11.84DD149 pKa = 3.37YY150 pKa = 10.92TYY152 pKa = 10.91TSLGTGTLNSTFFEE166 pKa = 4.37ATGTAEE172 pKa = 3.95IQKK175 pKa = 10.51ADD177 pKa = 3.25QTTWYY182 pKa = 9.71KK183 pKa = 11.03AIGLYY188 pKa = 10.41EE189 pKa = 4.21EE190 pKa = 5.69GYY192 pKa = 10.95DD193 pKa = 4.52IIFKK197 pKa = 10.13VAQDD201 pKa = 3.77GKK203 pKa = 9.07TVTVDD208 pKa = 3.27QQAAASDD215 pKa = 3.24ISGYY219 pKa = 8.37GTLYY223 pKa = 10.51VAGSGTLEE231 pKa = 3.97NGVITVEE238 pKa = 4.66LEE240 pKa = 4.08FTVSAGSFGSCTEE253 pKa = 3.87EE254 pKa = 5.47LILPAANN261 pKa = 3.27
Molecular weight: 27.41 kDa
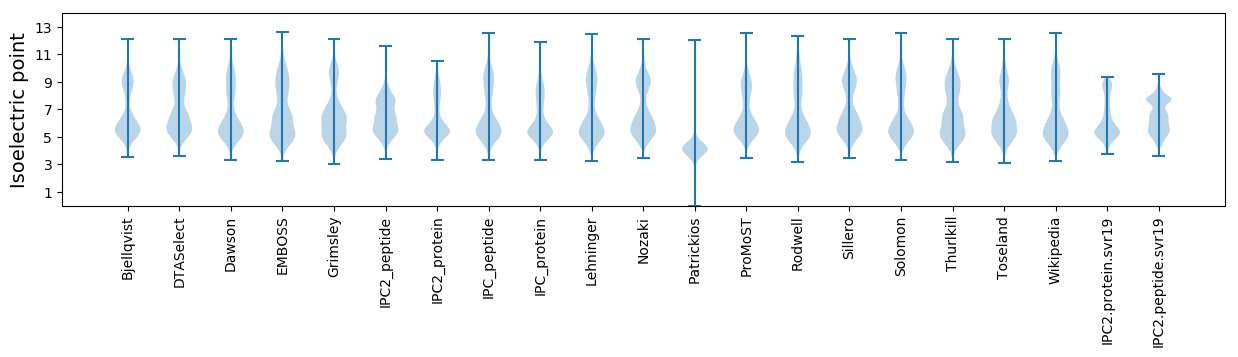
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6SLB9|R6SLB9_9BACE Uncharacterized protein OS=Bacteroides coprophilus CAG:333 OX=1263041 GN=BN612_00764 PE=4 SV=1
MM1 pKa = 7.35HH2 pKa = 6.65VRR4 pKa = 11.84HH5 pKa = 5.58TVEE8 pKa = 3.47MHH10 pKa = 4.77QVILAVINDD19 pKa = 3.94RR20 pKa = 11.84RR21 pKa = 11.84DD22 pKa = 3.38HH23 pKa = 6.56LPVLRR28 pKa = 11.84RR29 pKa = 11.84QTAEE33 pKa = 3.44QEE35 pKa = 3.72MRR37 pKa = 11.84IPVLFQIIRR46 pKa = 11.84HH47 pKa = 5.93LLRR50 pKa = 11.84LAQIRR55 pKa = 11.84TLLRR59 pKa = 11.84EE60 pKa = 3.68MPVRR64 pKa = 11.84FKK66 pKa = 10.96IEE68 pKa = 4.06IPQHH72 pKa = 4.81TVSCPALRR80 pKa = 11.84RR81 pKa = 11.84QFLIQLPVVVRR92 pKa = 11.84TAHH95 pKa = 6.09QYY97 pKa = 10.38HH98 pKa = 6.74IPQVTALRR106 pKa = 11.84TVRR109 pKa = 11.84LDD111 pKa = 3.06QHH113 pKa = 5.55TQAQPEE119 pKa = 4.2HH120 pKa = 6.77RR121 pKa = 11.84HH122 pKa = 5.24TDD124 pKa = 3.05QHH126 pKa = 5.68RR127 pKa = 11.84QQVRR131 pKa = 11.84QKK133 pKa = 8.12QLRR136 pKa = 11.84PGIHH140 pKa = 6.85ISSQLQDD147 pKa = 4.97SPAQDD152 pKa = 3.65CKK154 pKa = 9.84EE155 pKa = 3.91HH156 pKa = 6.97HH157 pKa = 6.85ILGHH161 pKa = 6.1TGQHH165 pKa = 7.51LIRR168 pKa = 11.84EE169 pKa = 4.52HH170 pKa = 6.49LPSVKK175 pKa = 10.13NYY177 pKa = 9.6PRR179 pKa = 11.84MHH181 pKa = 6.77RR182 pKa = 11.84TQQVTQHH189 pKa = 5.07QRR191 pKa = 11.84RR192 pKa = 11.84HH193 pKa = 4.78NLEE196 pKa = 4.15LRR198 pKa = 11.84CLEE201 pKa = 4.23CLLPGRR207 pKa = 11.84ILCPMIKK214 pKa = 9.75SVKK217 pKa = 9.5QSAYY221 pKa = 9.36DD222 pKa = 3.35TGIAGNDD229 pKa = 3.07KK230 pKa = 10.66CVNKK234 pKa = 10.04PVIRR238 pKa = 11.84LCNSSFHH245 pKa = 7.15DD246 pKa = 3.51LVLFNMFFYY255 pKa = 10.84KK256 pKa = 10.49KK257 pKa = 9.4FGYY260 pKa = 10.45FIFPLPRR267 pKa = 11.84EE268 pKa = 3.88LHH270 pKa = 6.36AFQLPITLLISRR282 pKa = 11.84II283 pKa = 3.86
MM1 pKa = 7.35HH2 pKa = 6.65VRR4 pKa = 11.84HH5 pKa = 5.58TVEE8 pKa = 3.47MHH10 pKa = 4.77QVILAVINDD19 pKa = 3.94RR20 pKa = 11.84RR21 pKa = 11.84DD22 pKa = 3.38HH23 pKa = 6.56LPVLRR28 pKa = 11.84RR29 pKa = 11.84QTAEE33 pKa = 3.44QEE35 pKa = 3.72MRR37 pKa = 11.84IPVLFQIIRR46 pKa = 11.84HH47 pKa = 5.93LLRR50 pKa = 11.84LAQIRR55 pKa = 11.84TLLRR59 pKa = 11.84EE60 pKa = 3.68MPVRR64 pKa = 11.84FKK66 pKa = 10.96IEE68 pKa = 4.06IPQHH72 pKa = 4.81TVSCPALRR80 pKa = 11.84RR81 pKa = 11.84QFLIQLPVVVRR92 pKa = 11.84TAHH95 pKa = 6.09QYY97 pKa = 10.38HH98 pKa = 6.74IPQVTALRR106 pKa = 11.84TVRR109 pKa = 11.84LDD111 pKa = 3.06QHH113 pKa = 5.55TQAQPEE119 pKa = 4.2HH120 pKa = 6.77RR121 pKa = 11.84HH122 pKa = 5.24TDD124 pKa = 3.05QHH126 pKa = 5.68RR127 pKa = 11.84QQVRR131 pKa = 11.84QKK133 pKa = 8.12QLRR136 pKa = 11.84PGIHH140 pKa = 6.85ISSQLQDD147 pKa = 4.97SPAQDD152 pKa = 3.65CKK154 pKa = 9.84EE155 pKa = 3.91HH156 pKa = 6.97HH157 pKa = 6.85ILGHH161 pKa = 6.1TGQHH165 pKa = 7.51LIRR168 pKa = 11.84EE169 pKa = 4.52HH170 pKa = 6.49LPSVKK175 pKa = 10.13NYY177 pKa = 9.6PRR179 pKa = 11.84MHH181 pKa = 6.77RR182 pKa = 11.84TQQVTQHH189 pKa = 5.07QRR191 pKa = 11.84RR192 pKa = 11.84HH193 pKa = 4.78NLEE196 pKa = 4.15LRR198 pKa = 11.84CLEE201 pKa = 4.23CLLPGRR207 pKa = 11.84ILCPMIKK214 pKa = 9.75SVKK217 pKa = 9.5QSAYY221 pKa = 9.36DD222 pKa = 3.35TGIAGNDD229 pKa = 3.07KK230 pKa = 10.66CVNKK234 pKa = 10.04PVIRR238 pKa = 11.84LCNSSFHH245 pKa = 7.15DD246 pKa = 3.51LVLFNMFFYY255 pKa = 10.84KK256 pKa = 10.49KK257 pKa = 9.4FGYY260 pKa = 10.45FIFPLPRR267 pKa = 11.84EE268 pKa = 3.88LHH270 pKa = 6.36AFQLPITLLISRR282 pKa = 11.84II283 pKa = 3.86
Molecular weight: 33.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
846506 |
30 |
1536 |
359.9 |
40.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.404 ± 0.044 | 1.343 ± 0.017 |
5.36 ± 0.03 | 6.696 ± 0.046 |
4.511 ± 0.037 | 7.043 ± 0.04 |
2.016 ± 0.021 | 6.476 ± 0.043 |
6.006 ± 0.036 | 9.557 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.82 ± 0.023 | 4.614 ± 0.043 |
3.953 ± 0.028 | 3.71 ± 0.03 |
5.031 ± 0.039 | 5.894 ± 0.039 |
5.434 ± 0.03 | 6.582 ± 0.044 |
1.278 ± 0.021 | 4.272 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
