
Rachiplusia ou multiple nucleopolyhedrovirus (strain R1) (RoMNPV)
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Baculoviridae; Alphabaculovirus; Autographa californica multiple nucleopolyhedrovirus; Rachiplusia ou MNPV
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
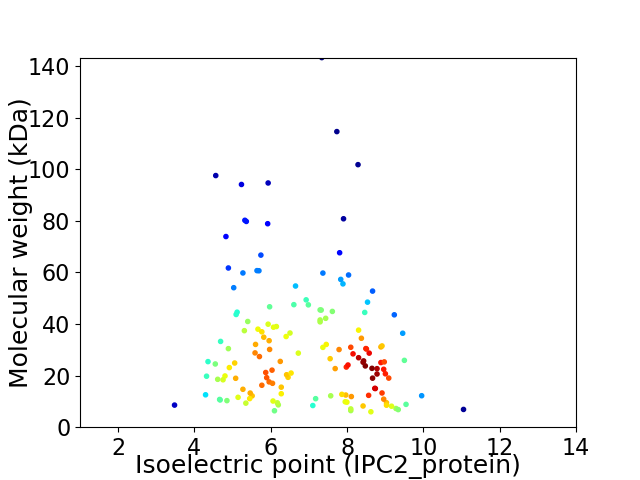
Virtual 2D-PAGE plot for 149 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8B9J8|Q8B9J8_NPVR1 Uncharacterized protein OS=Rachiplusia ou multiple nucleopolyhedrovirus (strain R1) OX=654904 PE=4 SV=1
MM1 pKa = 6.79TNAWFATDD9 pKa = 3.81VNLINCILKK18 pKa = 10.85DD19 pKa = 3.73NLFLVDD25 pKa = 3.73NNYY28 pKa = 10.24IILNVFDD35 pKa = 4.36QEE37 pKa = 4.17TDD39 pKa = 3.26QVRR42 pKa = 11.84PLCLGEE48 pKa = 4.24INALQTDD55 pKa = 4.11AAAQADD61 pKa = 3.89AMLDD65 pKa = 3.42TSSTSEE71 pKa = 4.07LQSNASTT78 pKa = 3.69
MM1 pKa = 6.79TNAWFATDD9 pKa = 3.81VNLINCILKK18 pKa = 10.85DD19 pKa = 3.73NLFLVDD25 pKa = 3.73NNYY28 pKa = 10.24IILNVFDD35 pKa = 4.36QEE37 pKa = 4.17TDD39 pKa = 3.26QVRR42 pKa = 11.84PLCLGEE48 pKa = 4.24INALQTDD55 pKa = 4.11AAAQADD61 pKa = 3.89AMLDD65 pKa = 3.42TSSTSEE71 pKa = 4.07LQSNASTT78 pKa = 3.69
Molecular weight: 8.57 kDa
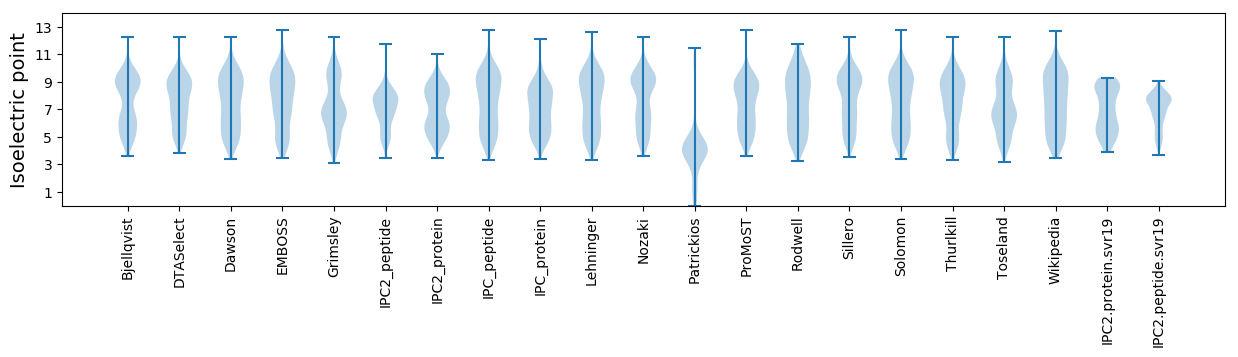
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8B9D2|Q8B9D2_NPVR1 Gp16 OS=Rachiplusia ou multiple nucleopolyhedrovirus (strain R1) OX=654904 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 8.07PTNNVMFDD10 pKa = 3.7DD11 pKa = 6.11ASVLWIDD18 pKa = 3.72TDD20 pKa = 4.41YY21 pKa = 11.02IYY23 pKa = 11.21QNLKK27 pKa = 9.57MPLQAFQQLLFTIPSKK43 pKa = 10.03HH44 pKa = 5.88RR45 pKa = 11.84KK46 pKa = 7.82MINDD50 pKa = 3.71AGGSCHH56 pKa = 5.81NTVKK60 pKa = 10.97YY61 pKa = 9.87MVDD64 pKa = 2.77IYY66 pKa = 11.18GAAVLVLRR74 pKa = 11.84TPCSFADD81 pKa = 3.49QLLSTFIANNYY92 pKa = 8.53LCYY95 pKa = 9.83FYY97 pKa = 10.79RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84SRR103 pKa = 11.84SRR105 pKa = 11.84SRR107 pKa = 11.84SRR109 pKa = 11.84SRR111 pKa = 11.84SPHH114 pKa = 5.16CRR116 pKa = 11.84PRR118 pKa = 11.84SRR120 pKa = 11.84SPHH123 pKa = 5.28CRR125 pKa = 11.84PRR127 pKa = 11.84SRR129 pKa = 11.84SRR131 pKa = 11.84SRR133 pKa = 11.84SRR135 pKa = 11.84SRR137 pKa = 11.84SRR139 pKa = 11.84SSSPRR144 pKa = 11.84RR145 pKa = 11.84GRR147 pKa = 11.84RR148 pKa = 11.84QIFEE152 pKa = 3.99ALEE155 pKa = 4.39KK156 pKa = 10.16IRR158 pKa = 11.84HH159 pKa = 5.1QNDD162 pKa = 2.9MLMSNVNQINLNQTNQFLEE181 pKa = 4.64LSNMMTGVRR190 pKa = 11.84NQNVQLLAALEE201 pKa = 4.14TAKK204 pKa = 10.87DD205 pKa = 3.83VILTRR210 pKa = 11.84LNTLLAEE217 pKa = 4.59ITDD220 pKa = 4.18SLPDD224 pKa = 3.73LTSMLDD230 pKa = 3.42KK231 pKa = 10.81LAEE234 pKa = 4.0QLLDD238 pKa = 4.78AINTVQQTLRR248 pKa = 11.84NEE250 pKa = 4.37LNNTNSILTNLASSVTNINGTLNNLLAAIEE280 pKa = 3.93NLIGGGGGGGNFNEE294 pKa = 5.08ADD296 pKa = 3.97RR297 pKa = 11.84QKK299 pKa = 11.49LDD301 pKa = 3.68LVYY304 pKa = 10.68TLVNEE309 pKa = 4.64IKK311 pKa = 10.78NILTGTLTKK320 pKa = 10.49KK321 pKa = 10.64
MM1 pKa = 7.57KK2 pKa = 8.07PTNNVMFDD10 pKa = 3.7DD11 pKa = 6.11ASVLWIDD18 pKa = 3.72TDD20 pKa = 4.41YY21 pKa = 11.02IYY23 pKa = 11.21QNLKK27 pKa = 9.57MPLQAFQQLLFTIPSKK43 pKa = 10.03HH44 pKa = 5.88RR45 pKa = 11.84KK46 pKa = 7.82MINDD50 pKa = 3.71AGGSCHH56 pKa = 5.81NTVKK60 pKa = 10.97YY61 pKa = 9.87MVDD64 pKa = 2.77IYY66 pKa = 11.18GAAVLVLRR74 pKa = 11.84TPCSFADD81 pKa = 3.49QLLSTFIANNYY92 pKa = 8.53LCYY95 pKa = 9.83FYY97 pKa = 10.79RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84SRR103 pKa = 11.84SRR105 pKa = 11.84SRR107 pKa = 11.84SRR109 pKa = 11.84SRR111 pKa = 11.84SPHH114 pKa = 5.16CRR116 pKa = 11.84PRR118 pKa = 11.84SRR120 pKa = 11.84SPHH123 pKa = 5.28CRR125 pKa = 11.84PRR127 pKa = 11.84SRR129 pKa = 11.84SRR131 pKa = 11.84SRR133 pKa = 11.84SRR135 pKa = 11.84SRR137 pKa = 11.84SRR139 pKa = 11.84SSSPRR144 pKa = 11.84RR145 pKa = 11.84GRR147 pKa = 11.84RR148 pKa = 11.84QIFEE152 pKa = 3.99ALEE155 pKa = 4.39KK156 pKa = 10.16IRR158 pKa = 11.84HH159 pKa = 5.1QNDD162 pKa = 2.9MLMSNVNQINLNQTNQFLEE181 pKa = 4.64LSNMMTGVRR190 pKa = 11.84NQNVQLLAALEE201 pKa = 4.14TAKK204 pKa = 10.87DD205 pKa = 3.83VILTRR210 pKa = 11.84LNTLLAEE217 pKa = 4.59ITDD220 pKa = 4.18SLPDD224 pKa = 3.73LTSMLDD230 pKa = 3.42KK231 pKa = 10.81LAEE234 pKa = 4.0QLLDD238 pKa = 4.78AINTVQQTLRR248 pKa = 11.84NEE250 pKa = 4.37LNNTNSILTNLASSVTNINGTLNNLLAAIEE280 pKa = 3.93NLIGGGGGGGNFNEE294 pKa = 5.08ADD296 pKa = 3.97RR297 pKa = 11.84QKK299 pKa = 11.49LDD301 pKa = 3.68LVYY304 pKa = 10.68TLVNEE309 pKa = 4.64IKK311 pKa = 10.78NILTGTLTKK320 pKa = 10.49KK321 pKa = 10.64
Molecular weight: 36.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
41291 |
53 |
1221 |
277.1 |
32.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.035 ± 0.175 | 2.56 ± 0.161 |
5.946 ± 0.152 | 5.253 ± 0.134 |
5.037 ± 0.134 | 3.238 ± 0.173 |
2.231 ± 0.11 | 6.788 ± 0.133 |
7.314 ± 0.271 | 9.399 ± 0.161 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.722 ± 0.078 | 8.179 ± 0.196 |
3.851 ± 0.26 | 3.768 ± 0.133 |
4.48 ± 0.169 | 6.285 ± 0.162 |
5.662 ± 0.151 | 6.488 ± 0.129 |
0.77 ± 0.069 | 4.994 ± 0.137 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
