
Firmicutes bacterium CAG:110
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
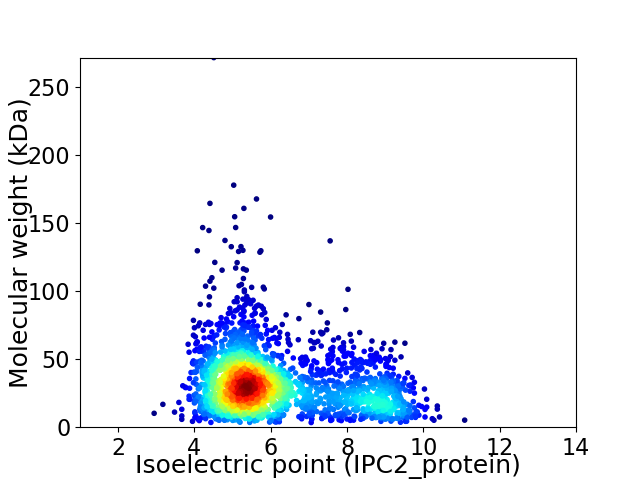
Virtual 2D-PAGE plot for 2311 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
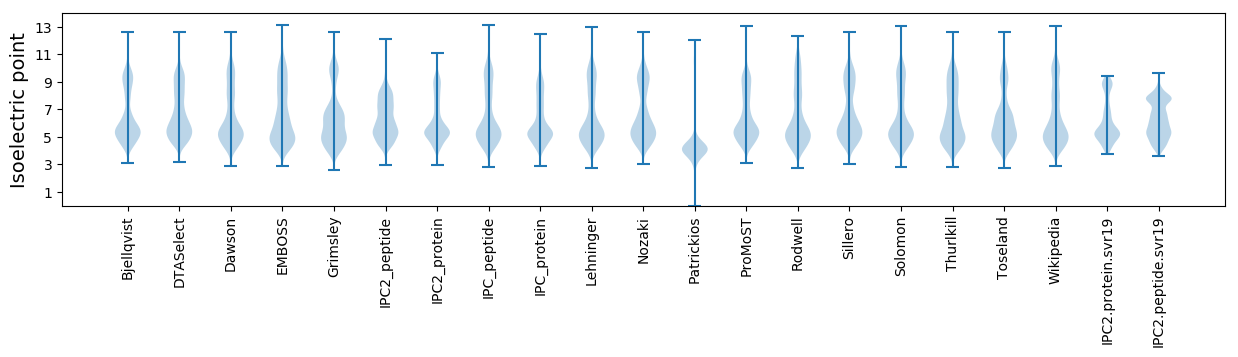
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5F079|R5F079_9FIRM Protein phosphatase 2C OS=Firmicutes bacterium CAG:110 OX=1263000 GN=BN466_00167 PE=4 SV=1
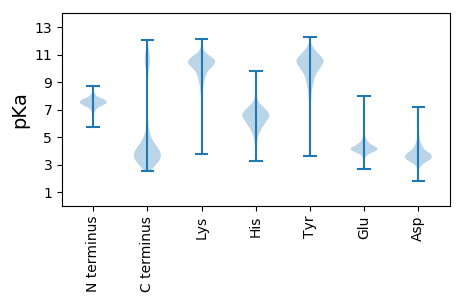
MM1 pKa = 7.45KK2 pKa = 10.45KK3 pKa = 10.26SLTRR7 pKa = 11.84VLLACLLVLTVVFACGCSNTPDD29 pKa = 3.67PSASNWKK36 pKa = 7.78VTFYY40 pKa = 10.74EE41 pKa = 4.59GDD43 pKa = 3.47GTTVLQEE50 pKa = 4.01VEE52 pKa = 4.41VADD55 pKa = 4.41GEE57 pKa = 4.46PVAMPEE63 pKa = 4.15LTKK66 pKa = 10.48EE67 pKa = 4.29GYY69 pKa = 9.93IIEE72 pKa = 5.15GYY74 pKa = 10.04YY75 pKa = 8.98ATPALKK81 pKa = 10.78VEE83 pKa = 4.59FDD85 pKa = 3.36VTQPITEE92 pKa = 4.07DD93 pKa = 3.14TAIFVAWQSSVVDD106 pKa = 3.7EE107 pKa = 4.92RR108 pKa = 11.84PWMLAGSLAGYY119 pKa = 8.87PDD121 pKa = 3.92NAWGKK126 pKa = 9.21IWPQNDD132 pKa = 3.39YY133 pKa = 11.04LLQPMEE139 pKa = 4.49GEE141 pKa = 4.34FNTFYY146 pKa = 10.79IDD148 pKa = 3.34VNLYY152 pKa = 10.73AGDD155 pKa = 3.72AFKK158 pKa = 10.66IAVIGEE164 pKa = 4.4GYY166 pKa = 10.5VWDD169 pKa = 4.37NDD171 pKa = 3.35NSIDD175 pKa = 4.91ASHH178 pKa = 7.33LANKK182 pKa = 8.98TEE184 pKa = 4.07SAVLCSGEE192 pKa = 3.9NAFDD196 pKa = 3.31SGANIEE202 pKa = 4.11VLEE205 pKa = 4.63DD206 pKa = 3.21GKK208 pKa = 11.31YY209 pKa = 10.33RR210 pKa = 11.84LILKK214 pKa = 8.98TDD216 pKa = 3.39AEE218 pKa = 4.48TLSLCSLSYY227 pKa = 10.75EE228 pKa = 4.19RR229 pKa = 11.84LGDD232 pKa = 3.88ADD234 pKa = 4.13EE235 pKa = 5.14KK236 pKa = 10.97PVAEE240 pKa = 5.04LSYY243 pKa = 11.62DD244 pKa = 3.49MKK246 pKa = 11.07LWASFNDD253 pKa = 3.54WSGQDD258 pKa = 3.37MQRR261 pKa = 11.84NGDD264 pKa = 4.4DD265 pKa = 4.57LIWWCEE271 pKa = 3.3ADD273 pKa = 3.8VPAGGGEE280 pKa = 3.98FGIKK284 pKa = 8.69NAAMDD289 pKa = 3.79GWYY292 pKa = 10.48SSEE295 pKa = 3.99NNSKK299 pKa = 11.06NIVLEE304 pKa = 4.2EE305 pKa = 3.67GHH307 pKa = 6.4YY308 pKa = 10.03MFFIEE313 pKa = 5.72LEE315 pKa = 4.39VVDD318 pKa = 5.19GKK320 pKa = 10.09PVLKK324 pKa = 10.77DD325 pKa = 3.32EE326 pKa = 5.36IIAEE330 pKa = 4.13EE331 pKa = 3.7PAYY334 pKa = 10.6YY335 pKa = 10.58VVGTCGNAGWAADD348 pKa = 3.59ANAEE352 pKa = 3.91NTAYY356 pKa = 10.09KK357 pKa = 8.37MTEE360 pKa = 3.67QDD362 pKa = 2.96SKK364 pKa = 11.82YY365 pKa = 10.25VLTVTFTEE373 pKa = 4.91DD374 pKa = 3.13EE375 pKa = 4.44TVDD378 pKa = 3.02WAEE381 pKa = 3.81NKK383 pKa = 9.76VAFKK387 pKa = 10.64VAYY390 pKa = 9.49GCGGKK395 pKa = 9.28VANEE399 pKa = 3.65NWFGAEE405 pKa = 3.79GMDD408 pKa = 4.62NVTVDD413 pKa = 3.44PGTYY417 pKa = 9.98TITFDD422 pKa = 3.79PATGAVTVEE431 pKa = 4.02
MM1 pKa = 7.45KK2 pKa = 10.45KK3 pKa = 10.26SLTRR7 pKa = 11.84VLLACLLVLTVVFACGCSNTPDD29 pKa = 3.67PSASNWKK36 pKa = 7.78VTFYY40 pKa = 10.74EE41 pKa = 4.59GDD43 pKa = 3.47GTTVLQEE50 pKa = 4.01VEE52 pKa = 4.41VADD55 pKa = 4.41GEE57 pKa = 4.46PVAMPEE63 pKa = 4.15LTKK66 pKa = 10.48EE67 pKa = 4.29GYY69 pKa = 9.93IIEE72 pKa = 5.15GYY74 pKa = 10.04YY75 pKa = 8.98ATPALKK81 pKa = 10.78VEE83 pKa = 4.59FDD85 pKa = 3.36VTQPITEE92 pKa = 4.07DD93 pKa = 3.14TAIFVAWQSSVVDD106 pKa = 3.7EE107 pKa = 4.92RR108 pKa = 11.84PWMLAGSLAGYY119 pKa = 8.87PDD121 pKa = 3.92NAWGKK126 pKa = 9.21IWPQNDD132 pKa = 3.39YY133 pKa = 11.04LLQPMEE139 pKa = 4.49GEE141 pKa = 4.34FNTFYY146 pKa = 10.79IDD148 pKa = 3.34VNLYY152 pKa = 10.73AGDD155 pKa = 3.72AFKK158 pKa = 10.66IAVIGEE164 pKa = 4.4GYY166 pKa = 10.5VWDD169 pKa = 4.37NDD171 pKa = 3.35NSIDD175 pKa = 4.91ASHH178 pKa = 7.33LANKK182 pKa = 8.98TEE184 pKa = 4.07SAVLCSGEE192 pKa = 3.9NAFDD196 pKa = 3.31SGANIEE202 pKa = 4.11VLEE205 pKa = 4.63DD206 pKa = 3.21GKK208 pKa = 11.31YY209 pKa = 10.33RR210 pKa = 11.84LILKK214 pKa = 8.98TDD216 pKa = 3.39AEE218 pKa = 4.48TLSLCSLSYY227 pKa = 10.75EE228 pKa = 4.19RR229 pKa = 11.84LGDD232 pKa = 3.88ADD234 pKa = 4.13EE235 pKa = 5.14KK236 pKa = 10.97PVAEE240 pKa = 5.04LSYY243 pKa = 11.62DD244 pKa = 3.49MKK246 pKa = 11.07LWASFNDD253 pKa = 3.54WSGQDD258 pKa = 3.37MQRR261 pKa = 11.84NGDD264 pKa = 4.4DD265 pKa = 4.57LIWWCEE271 pKa = 3.3ADD273 pKa = 3.8VPAGGGEE280 pKa = 3.98FGIKK284 pKa = 8.69NAAMDD289 pKa = 3.79GWYY292 pKa = 10.48SSEE295 pKa = 3.99NNSKK299 pKa = 11.06NIVLEE304 pKa = 4.2EE305 pKa = 3.67GHH307 pKa = 6.4YY308 pKa = 10.03MFFIEE313 pKa = 5.72LEE315 pKa = 4.39VVDD318 pKa = 5.19GKK320 pKa = 10.09PVLKK324 pKa = 10.77DD325 pKa = 3.32EE326 pKa = 5.36IIAEE330 pKa = 4.13EE331 pKa = 3.7PAYY334 pKa = 10.6YY335 pKa = 10.58VVGTCGNAGWAADD348 pKa = 3.59ANAEE352 pKa = 3.91NTAYY356 pKa = 10.09KK357 pKa = 8.37MTEE360 pKa = 3.67QDD362 pKa = 2.96SKK364 pKa = 11.82YY365 pKa = 10.25VLTVTFTEE373 pKa = 4.91DD374 pKa = 3.13EE375 pKa = 4.44TVDD378 pKa = 3.02WAEE381 pKa = 3.81NKK383 pKa = 9.76VAFKK387 pKa = 10.64VAYY390 pKa = 9.49GCGGKK395 pKa = 9.28VANEE399 pKa = 3.65NWFGAEE405 pKa = 3.79GMDD408 pKa = 4.62NVTVDD413 pKa = 3.44PGTYY417 pKa = 9.98TITFDD422 pKa = 3.79PATGAVTVEE431 pKa = 4.02
Molecular weight: 47.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5EHD0|R5EHD0_9FIRM Putative membrane protein insertion efficiency factor OS=Firmicutes bacterium CAG:110 OX=1263000 GN=BN466_01037 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.26QPSKK9 pKa = 8.09RR10 pKa = 11.84HH11 pKa = 5.53RR12 pKa = 11.84SMVHH16 pKa = 5.93GFRR19 pKa = 11.84QRR21 pKa = 11.84MATSNGRR28 pKa = 11.84KK29 pKa = 8.67VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.26QPSKK9 pKa = 8.09RR10 pKa = 11.84HH11 pKa = 5.53RR12 pKa = 11.84SMVHH16 pKa = 5.93GFRR19 pKa = 11.84QRR21 pKa = 11.84MATSNGRR28 pKa = 11.84KK29 pKa = 8.67VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
703987 |
30 |
2589 |
304.6 |
33.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.241 ± 0.049 | 1.761 ± 0.024 |
5.717 ± 0.044 | 6.471 ± 0.051 |
3.911 ± 0.038 | 7.815 ± 0.044 |
1.792 ± 0.025 | 5.94 ± 0.038 |
5.129 ± 0.049 | 9.558 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.028 | 3.625 ± 0.029 |
4.031 ± 0.034 | 3.259 ± 0.029 |
5.229 ± 0.048 | 5.571 ± 0.041 |
5.838 ± 0.055 | 7.353 ± 0.044 |
1.088 ± 0.019 | 3.731 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
