
Rotavirus X (isolate RVX/Human/Bangladesh/NADRV-B219/2002/GXP[X]) (RV ADRV-N) (Rotavirus (isolate novel adult diarrhea rotavirus-B219))
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; unclassified Rotavirus
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
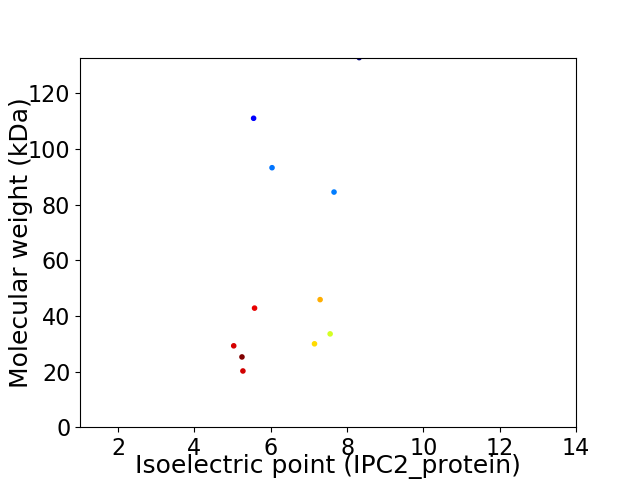
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q0H8C4|VP6_ROTB2 Intermediate capsid protein VP6 OS=Rotavirus X (isolate RVX/Human/Bangladesh/NADRV-B219/2002/GXP[X]) OX=348136 PE=3 SV=1
MM1 pKa = 7.7LLFIILSVGVDD12 pKa = 4.25AISYY16 pKa = 8.33IQNDD20 pKa = 3.93RR21 pKa = 11.84SNDD24 pKa = 3.08LCIIYY29 pKa = 10.54EE30 pKa = 4.15MTSFGASFNNANDD43 pKa = 3.83SLVKK47 pKa = 9.89LHH49 pKa = 6.32RR50 pKa = 11.84HH51 pKa = 5.3MGLKK55 pKa = 10.41YY56 pKa = 9.09EE57 pKa = 3.98ICKK60 pKa = 10.1IEE62 pKa = 4.45SNANALTQMQKK73 pKa = 10.68CNCLYY78 pKa = 10.81DD79 pKa = 3.86DD80 pKa = 4.17TPQIVVFTNFRR91 pKa = 11.84KK92 pKa = 10.21SSLKK96 pKa = 9.81TLIGTEE102 pKa = 4.27NKK104 pKa = 10.25CEE106 pKa = 4.4LLPQTTIYY114 pKa = 9.93TPTVDD119 pKa = 3.67IEE121 pKa = 4.43SEE123 pKa = 4.23YY124 pKa = 10.65FIYY127 pKa = 10.84GNDD130 pKa = 3.27VKK132 pKa = 10.65ICYY135 pKa = 9.55LDD137 pKa = 3.8KK138 pKa = 10.99NLLGIGCDD146 pKa = 3.65ATDD149 pKa = 3.49TTSWLDD155 pKa = 3.57LDD157 pKa = 5.21AGLPTNHH164 pKa = 7.35AIDD167 pKa = 3.75IPEE170 pKa = 3.97ITSDD174 pKa = 3.72GFKK177 pKa = 10.95LFAKK181 pKa = 10.45YY182 pKa = 10.4GDD184 pKa = 3.87SFLCQRR190 pKa = 11.84LMDD193 pKa = 4.14EE194 pKa = 4.83PKK196 pKa = 10.66KK197 pKa = 10.58QIQFYY202 pKa = 11.08AEE204 pKa = 3.92VDD206 pKa = 3.68NVPSNDD212 pKa = 3.39VIEE215 pKa = 4.87SSRR218 pKa = 11.84SWASVWKK225 pKa = 9.76VVKK228 pKa = 9.48TVLHH232 pKa = 5.47FTYY235 pKa = 10.35HH236 pKa = 6.84ILDD239 pKa = 3.99LFYY242 pKa = 11.11GNKK245 pKa = 9.52RR246 pKa = 11.84ATARR250 pKa = 11.84MIEE253 pKa = 4.08HH254 pKa = 6.6SPLGG258 pKa = 3.71
MM1 pKa = 7.7LLFIILSVGVDD12 pKa = 4.25AISYY16 pKa = 8.33IQNDD20 pKa = 3.93RR21 pKa = 11.84SNDD24 pKa = 3.08LCIIYY29 pKa = 10.54EE30 pKa = 4.15MTSFGASFNNANDD43 pKa = 3.83SLVKK47 pKa = 9.89LHH49 pKa = 6.32RR50 pKa = 11.84HH51 pKa = 5.3MGLKK55 pKa = 10.41YY56 pKa = 9.09EE57 pKa = 3.98ICKK60 pKa = 10.1IEE62 pKa = 4.45SNANALTQMQKK73 pKa = 10.68CNCLYY78 pKa = 10.81DD79 pKa = 3.86DD80 pKa = 4.17TPQIVVFTNFRR91 pKa = 11.84KK92 pKa = 10.21SSLKK96 pKa = 9.81TLIGTEE102 pKa = 4.27NKK104 pKa = 10.25CEE106 pKa = 4.4LLPQTTIYY114 pKa = 9.93TPTVDD119 pKa = 3.67IEE121 pKa = 4.43SEE123 pKa = 4.23YY124 pKa = 10.65FIYY127 pKa = 10.84GNDD130 pKa = 3.27VKK132 pKa = 10.65ICYY135 pKa = 9.55LDD137 pKa = 3.8KK138 pKa = 10.99NLLGIGCDD146 pKa = 3.65ATDD149 pKa = 3.49TTSWLDD155 pKa = 3.57LDD157 pKa = 5.21AGLPTNHH164 pKa = 7.35AIDD167 pKa = 3.75IPEE170 pKa = 3.97ITSDD174 pKa = 3.72GFKK177 pKa = 10.95LFAKK181 pKa = 10.45YY182 pKa = 10.4GDD184 pKa = 3.87SFLCQRR190 pKa = 11.84LMDD193 pKa = 4.14EE194 pKa = 4.83PKK196 pKa = 10.66KK197 pKa = 10.58QIQFYY202 pKa = 11.08AEE204 pKa = 3.92VDD206 pKa = 3.68NVPSNDD212 pKa = 3.39VIEE215 pKa = 4.87SSRR218 pKa = 11.84SWASVWKK225 pKa = 9.76VVKK228 pKa = 9.48TVLHH232 pKa = 5.47FTYY235 pKa = 10.35HH236 pKa = 6.84ILDD239 pKa = 3.99LFYY242 pKa = 11.11GNKK245 pKa = 9.52RR246 pKa = 11.84ATARR250 pKa = 11.84MIEE253 pKa = 4.08HH254 pKa = 6.6SPLGG258 pKa = 3.71
Molecular weight: 29.26 kDa
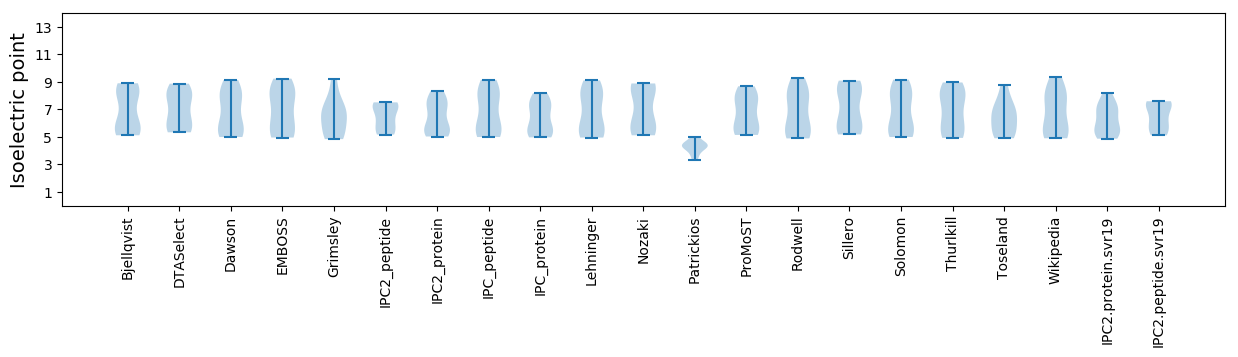
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A9Q1K8|VP2_ROTB2 Inner capsid protein VP2 OS=Rotavirus X (isolate RVX/Human/Bangladesh/NADRV-B219/2002/GXP[X]) OX=348136 PE=2 SV=1
MM1 pKa = 7.46EE2 pKa = 5.77PEE4 pKa = 5.3NYY6 pKa = 9.97LAWLARR12 pKa = 11.84DD13 pKa = 3.39IVRR16 pKa = 11.84NLSYY20 pKa = 10.97TSLVYY25 pKa = 10.81NNPKK29 pKa = 9.55VAIVEE34 pKa = 4.04LLDD37 pKa = 3.62NKK39 pKa = 10.42EE40 pKa = 4.02AFFTYY45 pKa = 9.81EE46 pKa = 4.31KK47 pKa = 10.21EE48 pKa = 4.28QKK50 pKa = 9.33TPEE53 pKa = 3.84ALINYY58 pKa = 8.46IDD60 pKa = 4.83SIVKK64 pKa = 10.34SSISVEE70 pKa = 4.21DD71 pKa = 4.19KK72 pKa = 11.0IEE74 pKa = 4.01ALLKK78 pKa = 9.63IRR80 pKa = 11.84YY81 pKa = 8.73ISVYY85 pKa = 10.67VDD87 pKa = 3.48DD88 pKa = 5.96KK89 pKa = 11.35SDD91 pKa = 3.21KK92 pKa = 10.6RR93 pKa = 11.84DD94 pKa = 3.24IVLQLLNRR102 pKa = 11.84TIKK105 pKa = 10.59KK106 pKa = 9.73IEE108 pKa = 4.23SKK110 pKa = 9.83TKK112 pKa = 10.19ISNEE116 pKa = 3.59LNDD119 pKa = 5.39AINAITIEE127 pKa = 3.99SRR129 pKa = 11.84NWKK132 pKa = 8.48IQNSEE137 pKa = 3.88SFKK140 pKa = 10.21PYY142 pKa = 10.12HH143 pKa = 5.95YY144 pKa = 10.24NQLVSDD150 pKa = 4.0FLKK153 pKa = 10.95YY154 pKa = 10.82NEE156 pKa = 4.06FEE158 pKa = 4.21ILEE161 pKa = 4.47GTDD164 pKa = 3.39PLKK167 pKa = 10.62WKK169 pKa = 10.67SDD171 pKa = 3.61TLQGLSPNYY180 pKa = 9.03NHH182 pKa = 6.95RR183 pKa = 11.84THH185 pKa = 6.5TLISSIIYY193 pKa = 7.14ATSVRR198 pKa = 11.84FDD200 pKa = 3.58NYY202 pKa = 10.75NDD204 pKa = 3.49EE205 pKa = 4.15QLQVLLYY212 pKa = 9.9LFSIIKK218 pKa = 8.36TNYY221 pKa = 7.69VNGYY225 pKa = 10.14LEE227 pKa = 4.16ILPNRR232 pKa = 11.84KK233 pKa = 8.51WSHH236 pKa = 5.57SLADD240 pKa = 3.54LRR242 pKa = 11.84EE243 pKa = 4.27NKK245 pKa = 10.34SIMMYY250 pKa = 8.9SAKK253 pKa = 9.91IIHH256 pKa = 7.0ASCAMISILHH266 pKa = 6.46AVPIDD271 pKa = 3.7YY272 pKa = 10.59FFLAQIIASFSEE284 pKa = 4.52IPAHH288 pKa = 6.57AAKK291 pKa = 10.27QLSSPMTLYY300 pKa = 10.33IGIAQLRR307 pKa = 11.84SNIVVSTKK315 pKa = 9.92IAAEE319 pKa = 4.22SVATEE324 pKa = 4.1SPNISRR330 pKa = 11.84LEE332 pKa = 3.9EE333 pKa = 3.92SQLRR337 pKa = 11.84EE338 pKa = 3.85WEE340 pKa = 4.13QEE342 pKa = 3.75MNEE345 pKa = 4.11YY346 pKa = 10.06PFQSSRR352 pKa = 11.84MVRR355 pKa = 11.84MMKK358 pKa = 10.45KK359 pKa = 10.16NIFDD363 pKa = 3.54VSVDD367 pKa = 3.17VFYY370 pKa = 11.01AIFNCFSATFHH381 pKa = 5.9VGHH384 pKa = 7.3RR385 pKa = 11.84IDD387 pKa = 4.2NPQDD391 pKa = 3.85AIEE394 pKa = 4.35AQVKK398 pKa = 9.37VEE400 pKa = 3.99YY401 pKa = 10.21TSDD404 pKa = 3.39VDD406 pKa = 4.52KK407 pKa = 11.43EE408 pKa = 4.44MYY410 pKa = 9.39DD411 pKa = 3.01QYY413 pKa = 11.88YY414 pKa = 10.5FLLKK418 pKa = 10.67RR419 pKa = 11.84MLTDD423 pKa = 3.02QLAEE427 pKa = 4.24YY428 pKa = 10.3AEE430 pKa = 4.03EE431 pKa = 4.9MYY433 pKa = 10.57FKK435 pKa = 11.09YY436 pKa = 10.85NSDD439 pKa = 3.22VTAEE443 pKa = 4.22SLAAMANSSNGYY455 pKa = 8.57SRR457 pKa = 11.84SVTFIDD463 pKa = 5.25RR464 pKa = 11.84EE465 pKa = 4.03IKK467 pKa = 7.04TTKK470 pKa = 10.6KK471 pKa = 9.58MLHH474 pKa = 6.67LDD476 pKa = 3.84DD477 pKa = 6.22DD478 pKa = 4.59LSKK481 pKa = 11.26NLNFTNIGEE490 pKa = 4.3QIKK493 pKa = 10.56KK494 pKa = 9.26GIPMGTRR501 pKa = 11.84NVPARR506 pKa = 11.84QTRR509 pKa = 11.84GIFILSWQVAAIQHH523 pKa = 6.29TIAEE527 pKa = 4.27FLYY530 pKa = 10.6KK531 pKa = 9.96KK532 pKa = 10.11AKK534 pKa = 9.99KK535 pKa = 10.15GGFGATFAEE544 pKa = 4.95AYY546 pKa = 9.22VSKK549 pKa = 10.69AATLTYY555 pKa = 10.47GILAEE560 pKa = 4.34ATSKK564 pKa = 11.11ADD566 pKa = 3.65QLILYY571 pKa = 8.59TDD573 pKa = 3.83VSQWDD578 pKa = 3.7ASQHH582 pKa = 4.09NTEE585 pKa = 4.89PYY587 pKa = 9.38RR588 pKa = 11.84SAWINAIKK596 pKa = 10.08EE597 pKa = 3.85ARR599 pKa = 11.84TKK601 pKa = 11.14YY602 pKa = 9.99KK603 pKa = 10.28INYY606 pKa = 7.3NQEE609 pKa = 3.97PVVLGMNVLDD619 pKa = 5.15KK620 pKa = 10.67MIEE623 pKa = 3.92IQEE626 pKa = 4.22ALLNSNLIVEE636 pKa = 4.62SQGSKK641 pKa = 9.77RR642 pKa = 11.84QPLRR646 pKa = 11.84IKK648 pKa = 9.73YY649 pKa = 9.41HH650 pKa = 5.48GVASGEE656 pKa = 4.15KK657 pKa = 6.55TTKK660 pKa = 10.0IGNSFANVALITTVFNNLTNTMPSIRR686 pKa = 11.84VNHH689 pKa = 5.67MRR691 pKa = 11.84VDD693 pKa = 3.56GDD695 pKa = 4.26DD696 pKa = 3.95NVVTMYY702 pKa = 8.46TANRR706 pKa = 11.84IDD708 pKa = 3.69EE709 pKa = 4.2VQEE712 pKa = 4.15NIKK715 pKa = 10.47EE716 pKa = 4.08KK717 pKa = 9.82YY718 pKa = 9.85KK719 pKa = 11.03RR720 pKa = 11.84MNAKK724 pKa = 9.63VKK726 pKa = 10.64ALASYY731 pKa = 9.47TGLEE735 pKa = 3.81MAKK738 pKa = 10.23RR739 pKa = 11.84FIICGKK745 pKa = 8.84IFEE748 pKa = 5.12RR749 pKa = 11.84GAISIFTAEE758 pKa = 4.28RR759 pKa = 11.84PYY761 pKa = 10.7GTDD764 pKa = 3.38LSVQSTTGSLIYY776 pKa = 10.34SAAVNAYY783 pKa = 9.89RR784 pKa = 11.84GFGDD788 pKa = 4.5DD789 pKa = 4.13YY790 pKa = 11.23LNFMTDD796 pKa = 3.01VLVPPSASVKK806 pKa = 8.23ITGRR810 pKa = 11.84LRR812 pKa = 11.84SLLSPVTLYY821 pKa = 9.93STGPLSFEE829 pKa = 3.53ITPYY833 pKa = 10.94GLGGRR838 pKa = 11.84MRR840 pKa = 11.84LFSLSKK846 pKa = 11.03EE847 pKa = 3.83NMEE850 pKa = 4.64LYY852 pKa = 10.53KK853 pKa = 10.48ILTSSLAISIQPDD866 pKa = 3.65EE867 pKa = 4.2IKK869 pKa = 10.39KK870 pKa = 10.68YY871 pKa = 10.93SSTPQFKK878 pKa = 10.71ARR880 pKa = 11.84VDD882 pKa = 3.6RR883 pKa = 11.84MISSVQIAMKK893 pKa = 10.45SEE895 pKa = 3.81AKK897 pKa = 10.19IITSILRR904 pKa = 11.84DD905 pKa = 3.5KK906 pKa = 11.2EE907 pKa = 4.15EE908 pKa = 3.97QKK910 pKa = 9.79TLGVPNVATAKK921 pKa = 10.12NRR923 pKa = 11.84QQIDD927 pKa = 3.45KK928 pKa = 10.73ARR930 pKa = 11.84KK931 pKa = 6.86TLSLPKK937 pKa = 10.17EE938 pKa = 3.96ILPKK942 pKa = 8.04VTKK945 pKa = 10.09YY946 pKa = 9.51YY947 pKa = 10.06PEE949 pKa = 4.86EE950 pKa = 4.0IFHH953 pKa = 7.04LILRR957 pKa = 11.84NSTLTIPKK965 pKa = 9.46LNTMTKK971 pKa = 9.78VYY973 pKa = 10.0MNNSVNITKK982 pKa = 9.84LQQQIGVRR990 pKa = 11.84VSSGIQVHH998 pKa = 6.09KK999 pKa = 10.27PINTLLKK1006 pKa = 10.35LVEE1009 pKa = 4.09KK1010 pKa = 10.25HH1011 pKa = 6.21SPIKK1015 pKa = 10.36ISPSDD1020 pKa = 4.09LILYY1024 pKa = 8.06SKK1026 pKa = 10.78KK1027 pKa = 10.5YY1028 pKa = 10.86DD1029 pKa = 3.53LTNLNGKK1036 pKa = 8.56KK1037 pKa = 9.65QFLMDD1042 pKa = 4.54LGISGNEE1049 pKa = 3.27LRR1051 pKa = 11.84FYY1053 pKa = 11.2LNSKK1057 pKa = 10.15LLFHH1061 pKa = 7.52DD1062 pKa = 5.02LLLSKK1067 pKa = 10.61YY1068 pKa = 9.83DD1069 pKa = 3.9KK1070 pKa = 10.88LYY1072 pKa = 9.48EE1073 pKa = 4.29APGFGATQLNALPLDD1088 pKa = 3.78LTAAEE1093 pKa = 4.59KK1094 pKa = 10.98VFSIKK1099 pKa = 10.75LNLPNTYY1106 pKa = 10.28YY1107 pKa = 10.79EE1108 pKa = 4.55LLMLVLLYY1116 pKa = 10.54EE1117 pKa = 4.22YY1118 pKa = 11.32VNFVMFTGNTFRR1130 pKa = 11.84AVCIPEE1136 pKa = 4.14SQTINAKK1143 pKa = 9.54LVKK1146 pKa = 9.71TIMTMIDD1153 pKa = 3.93NIQLDD1158 pKa = 4.0TVMFSDD1164 pKa = 5.32NIFF1167 pKa = 3.37
MM1 pKa = 7.46EE2 pKa = 5.77PEE4 pKa = 5.3NYY6 pKa = 9.97LAWLARR12 pKa = 11.84DD13 pKa = 3.39IVRR16 pKa = 11.84NLSYY20 pKa = 10.97TSLVYY25 pKa = 10.81NNPKK29 pKa = 9.55VAIVEE34 pKa = 4.04LLDD37 pKa = 3.62NKK39 pKa = 10.42EE40 pKa = 4.02AFFTYY45 pKa = 9.81EE46 pKa = 4.31KK47 pKa = 10.21EE48 pKa = 4.28QKK50 pKa = 9.33TPEE53 pKa = 3.84ALINYY58 pKa = 8.46IDD60 pKa = 4.83SIVKK64 pKa = 10.34SSISVEE70 pKa = 4.21DD71 pKa = 4.19KK72 pKa = 11.0IEE74 pKa = 4.01ALLKK78 pKa = 9.63IRR80 pKa = 11.84YY81 pKa = 8.73ISVYY85 pKa = 10.67VDD87 pKa = 3.48DD88 pKa = 5.96KK89 pKa = 11.35SDD91 pKa = 3.21KK92 pKa = 10.6RR93 pKa = 11.84DD94 pKa = 3.24IVLQLLNRR102 pKa = 11.84TIKK105 pKa = 10.59KK106 pKa = 9.73IEE108 pKa = 4.23SKK110 pKa = 9.83TKK112 pKa = 10.19ISNEE116 pKa = 3.59LNDD119 pKa = 5.39AINAITIEE127 pKa = 3.99SRR129 pKa = 11.84NWKK132 pKa = 8.48IQNSEE137 pKa = 3.88SFKK140 pKa = 10.21PYY142 pKa = 10.12HH143 pKa = 5.95YY144 pKa = 10.24NQLVSDD150 pKa = 4.0FLKK153 pKa = 10.95YY154 pKa = 10.82NEE156 pKa = 4.06FEE158 pKa = 4.21ILEE161 pKa = 4.47GTDD164 pKa = 3.39PLKK167 pKa = 10.62WKK169 pKa = 10.67SDD171 pKa = 3.61TLQGLSPNYY180 pKa = 9.03NHH182 pKa = 6.95RR183 pKa = 11.84THH185 pKa = 6.5TLISSIIYY193 pKa = 7.14ATSVRR198 pKa = 11.84FDD200 pKa = 3.58NYY202 pKa = 10.75NDD204 pKa = 3.49EE205 pKa = 4.15QLQVLLYY212 pKa = 9.9LFSIIKK218 pKa = 8.36TNYY221 pKa = 7.69VNGYY225 pKa = 10.14LEE227 pKa = 4.16ILPNRR232 pKa = 11.84KK233 pKa = 8.51WSHH236 pKa = 5.57SLADD240 pKa = 3.54LRR242 pKa = 11.84EE243 pKa = 4.27NKK245 pKa = 10.34SIMMYY250 pKa = 8.9SAKK253 pKa = 9.91IIHH256 pKa = 7.0ASCAMISILHH266 pKa = 6.46AVPIDD271 pKa = 3.7YY272 pKa = 10.59FFLAQIIASFSEE284 pKa = 4.52IPAHH288 pKa = 6.57AAKK291 pKa = 10.27QLSSPMTLYY300 pKa = 10.33IGIAQLRR307 pKa = 11.84SNIVVSTKK315 pKa = 9.92IAAEE319 pKa = 4.22SVATEE324 pKa = 4.1SPNISRR330 pKa = 11.84LEE332 pKa = 3.9EE333 pKa = 3.92SQLRR337 pKa = 11.84EE338 pKa = 3.85WEE340 pKa = 4.13QEE342 pKa = 3.75MNEE345 pKa = 4.11YY346 pKa = 10.06PFQSSRR352 pKa = 11.84MVRR355 pKa = 11.84MMKK358 pKa = 10.45KK359 pKa = 10.16NIFDD363 pKa = 3.54VSVDD367 pKa = 3.17VFYY370 pKa = 11.01AIFNCFSATFHH381 pKa = 5.9VGHH384 pKa = 7.3RR385 pKa = 11.84IDD387 pKa = 4.2NPQDD391 pKa = 3.85AIEE394 pKa = 4.35AQVKK398 pKa = 9.37VEE400 pKa = 3.99YY401 pKa = 10.21TSDD404 pKa = 3.39VDD406 pKa = 4.52KK407 pKa = 11.43EE408 pKa = 4.44MYY410 pKa = 9.39DD411 pKa = 3.01QYY413 pKa = 11.88YY414 pKa = 10.5FLLKK418 pKa = 10.67RR419 pKa = 11.84MLTDD423 pKa = 3.02QLAEE427 pKa = 4.24YY428 pKa = 10.3AEE430 pKa = 4.03EE431 pKa = 4.9MYY433 pKa = 10.57FKK435 pKa = 11.09YY436 pKa = 10.85NSDD439 pKa = 3.22VTAEE443 pKa = 4.22SLAAMANSSNGYY455 pKa = 8.57SRR457 pKa = 11.84SVTFIDD463 pKa = 5.25RR464 pKa = 11.84EE465 pKa = 4.03IKK467 pKa = 7.04TTKK470 pKa = 10.6KK471 pKa = 9.58MLHH474 pKa = 6.67LDD476 pKa = 3.84DD477 pKa = 6.22DD478 pKa = 4.59LSKK481 pKa = 11.26NLNFTNIGEE490 pKa = 4.3QIKK493 pKa = 10.56KK494 pKa = 9.26GIPMGTRR501 pKa = 11.84NVPARR506 pKa = 11.84QTRR509 pKa = 11.84GIFILSWQVAAIQHH523 pKa = 6.29TIAEE527 pKa = 4.27FLYY530 pKa = 10.6KK531 pKa = 9.96KK532 pKa = 10.11AKK534 pKa = 9.99KK535 pKa = 10.15GGFGATFAEE544 pKa = 4.95AYY546 pKa = 9.22VSKK549 pKa = 10.69AATLTYY555 pKa = 10.47GILAEE560 pKa = 4.34ATSKK564 pKa = 11.11ADD566 pKa = 3.65QLILYY571 pKa = 8.59TDD573 pKa = 3.83VSQWDD578 pKa = 3.7ASQHH582 pKa = 4.09NTEE585 pKa = 4.89PYY587 pKa = 9.38RR588 pKa = 11.84SAWINAIKK596 pKa = 10.08EE597 pKa = 3.85ARR599 pKa = 11.84TKK601 pKa = 11.14YY602 pKa = 9.99KK603 pKa = 10.28INYY606 pKa = 7.3NQEE609 pKa = 3.97PVVLGMNVLDD619 pKa = 5.15KK620 pKa = 10.67MIEE623 pKa = 3.92IQEE626 pKa = 4.22ALLNSNLIVEE636 pKa = 4.62SQGSKK641 pKa = 9.77RR642 pKa = 11.84QPLRR646 pKa = 11.84IKK648 pKa = 9.73YY649 pKa = 9.41HH650 pKa = 5.48GVASGEE656 pKa = 4.15KK657 pKa = 6.55TTKK660 pKa = 10.0IGNSFANVALITTVFNNLTNTMPSIRR686 pKa = 11.84VNHH689 pKa = 5.67MRR691 pKa = 11.84VDD693 pKa = 3.56GDD695 pKa = 4.26DD696 pKa = 3.95NVVTMYY702 pKa = 8.46TANRR706 pKa = 11.84IDD708 pKa = 3.69EE709 pKa = 4.2VQEE712 pKa = 4.15NIKK715 pKa = 10.47EE716 pKa = 4.08KK717 pKa = 9.82YY718 pKa = 9.85KK719 pKa = 11.03RR720 pKa = 11.84MNAKK724 pKa = 9.63VKK726 pKa = 10.64ALASYY731 pKa = 9.47TGLEE735 pKa = 3.81MAKK738 pKa = 10.23RR739 pKa = 11.84FIICGKK745 pKa = 8.84IFEE748 pKa = 5.12RR749 pKa = 11.84GAISIFTAEE758 pKa = 4.28RR759 pKa = 11.84PYY761 pKa = 10.7GTDD764 pKa = 3.38LSVQSTTGSLIYY776 pKa = 10.34SAAVNAYY783 pKa = 9.89RR784 pKa = 11.84GFGDD788 pKa = 4.5DD789 pKa = 4.13YY790 pKa = 11.23LNFMTDD796 pKa = 3.01VLVPPSASVKK806 pKa = 8.23ITGRR810 pKa = 11.84LRR812 pKa = 11.84SLLSPVTLYY821 pKa = 9.93STGPLSFEE829 pKa = 3.53ITPYY833 pKa = 10.94GLGGRR838 pKa = 11.84MRR840 pKa = 11.84LFSLSKK846 pKa = 11.03EE847 pKa = 3.83NMEE850 pKa = 4.64LYY852 pKa = 10.53KK853 pKa = 10.48ILTSSLAISIQPDD866 pKa = 3.65EE867 pKa = 4.2IKK869 pKa = 10.39KK870 pKa = 10.68YY871 pKa = 10.93SSTPQFKK878 pKa = 10.71ARR880 pKa = 11.84VDD882 pKa = 3.6RR883 pKa = 11.84MISSVQIAMKK893 pKa = 10.45SEE895 pKa = 3.81AKK897 pKa = 10.19IITSILRR904 pKa = 11.84DD905 pKa = 3.5KK906 pKa = 11.2EE907 pKa = 4.15EE908 pKa = 3.97QKK910 pKa = 9.79TLGVPNVATAKK921 pKa = 10.12NRR923 pKa = 11.84QQIDD927 pKa = 3.45KK928 pKa = 10.73ARR930 pKa = 11.84KK931 pKa = 6.86TLSLPKK937 pKa = 10.17EE938 pKa = 3.96ILPKK942 pKa = 8.04VTKK945 pKa = 10.09YY946 pKa = 9.51YY947 pKa = 10.06PEE949 pKa = 4.86EE950 pKa = 4.0IFHH953 pKa = 7.04LILRR957 pKa = 11.84NSTLTIPKK965 pKa = 9.46LNTMTKK971 pKa = 9.78VYY973 pKa = 10.0MNNSVNITKK982 pKa = 9.84LQQQIGVRR990 pKa = 11.84VSSGIQVHH998 pKa = 6.09KK999 pKa = 10.27PINTLLKK1006 pKa = 10.35LVEE1009 pKa = 4.09KK1010 pKa = 10.25HH1011 pKa = 6.21SPIKK1015 pKa = 10.36ISPSDD1020 pKa = 4.09LILYY1024 pKa = 8.06SKK1026 pKa = 10.78KK1027 pKa = 10.5YY1028 pKa = 10.86DD1029 pKa = 3.53LTNLNGKK1036 pKa = 8.56KK1037 pKa = 9.65QFLMDD1042 pKa = 4.54LGISGNEE1049 pKa = 3.27LRR1051 pKa = 11.84FYY1053 pKa = 11.2LNSKK1057 pKa = 10.15LLFHH1061 pKa = 7.52DD1062 pKa = 5.02LLLSKK1067 pKa = 10.61YY1068 pKa = 9.83DD1069 pKa = 3.9KK1070 pKa = 10.88LYY1072 pKa = 9.48EE1073 pKa = 4.29APGFGATQLNALPLDD1088 pKa = 3.78LTAAEE1093 pKa = 4.59KK1094 pKa = 10.98VFSIKK1099 pKa = 10.75LNLPNTYY1106 pKa = 10.28YY1107 pKa = 10.79EE1108 pKa = 4.55LLMLVLLYY1116 pKa = 10.54EE1117 pKa = 4.22YY1118 pKa = 11.32VNFVMFTGNTFRR1130 pKa = 11.84AVCIPEE1136 pKa = 4.14SQTINAKK1143 pKa = 9.54LVKK1146 pKa = 9.71TIMTMIDD1153 pKa = 3.93NIQLDD1158 pKa = 4.0TVMFSDD1164 pKa = 5.32NIFF1167 pKa = 3.37
Molecular weight: 132.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5682 |
176 |
1167 |
516.5 |
58.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.72 ± 0.615 | 1.197 ± 0.357 |
6.037 ± 0.526 | 5.949 ± 0.428 |
4.576 ± 0.331 | 3.995 ± 0.433 |
1.83 ± 0.244 | 8.624 ± 0.422 |
6.793 ± 0.511 | 8.659 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.182 | 6.829 ± 0.296 |
3.678 ± 0.34 | 3.96 ± 0.265 |
5.016 ± 0.454 | 7.744 ± 0.308 |
6.406 ± 0.433 | 5.544 ± 0.438 |
0.792 ± 0.083 | 3.907 ± 0.447 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
