
Chelatococcus asaccharovorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Chelatococcaceae; Chelatococcus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
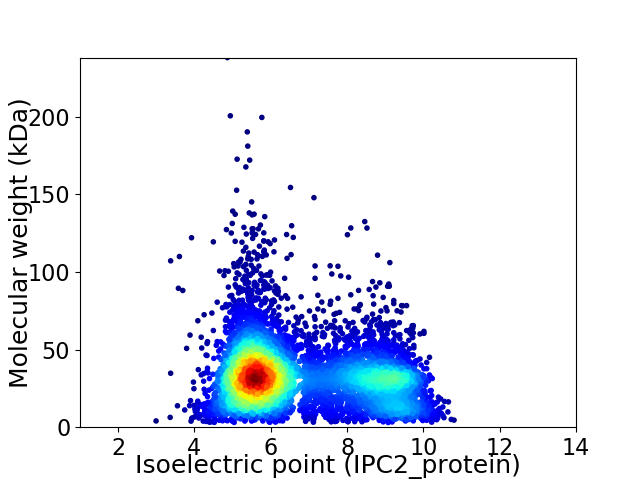
Virtual 2D-PAGE plot for 6659 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V3TT05|A0A2V3TT05_9RHIZ Uncharacterized protein OS=Chelatococcus asaccharovorans OX=28210 GN=C7450_1214 PE=4 SV=1
MM1 pKa = 7.44ATTDD5 pKa = 3.63AASSSLIPAHH15 pKa = 6.81SPFSYY20 pKa = 10.73NVGINYY26 pKa = 7.91EE27 pKa = 4.08SWDD30 pKa = 3.51GGRR33 pKa = 11.84VGRR36 pKa = 11.84VIANDD41 pKa = 3.69LDD43 pKa = 5.89AITQNFKK50 pKa = 11.09LIRR53 pKa = 11.84TYY55 pKa = 11.15HH56 pKa = 6.48DD57 pKa = 3.43AAVGTSDD64 pKa = 3.21PTAPIIEE71 pKa = 4.31PTQQEE76 pKa = 4.46VISYY80 pKa = 9.26VLAHH84 pKa = 6.99PGLEE88 pKa = 3.99LSMGTNNNALANGGYY103 pKa = 8.93GAPWSPGLMVDD114 pKa = 5.25PAYY117 pKa = 9.84TDD119 pKa = 2.85QWVAMIIEE127 pKa = 4.75AFGSTNAVEE136 pKa = 5.02EE137 pKa = 4.08NLKK140 pKa = 10.18MILLGNEE147 pKa = 3.83LDD149 pKa = 4.51ANGPPPGDD157 pKa = 3.51PLFEE161 pKa = 4.26TYY163 pKa = 10.76YY164 pKa = 10.85SVWIPTAFQNLQTSLANAGLGSIPISTTIANYY196 pKa = 8.14GTGNVVSTSISSFIEE211 pKa = 4.55TNWSASWNGGEE222 pKa = 5.26PIVLYY227 pKa = 10.51NQYY230 pKa = 9.2TPDD233 pKa = 3.72NGSSTDD239 pKa = 3.51YY240 pKa = 11.1SSVISYY246 pKa = 10.08FEE248 pKa = 4.12SLATTFDD255 pKa = 3.3GALAPFIGEE264 pKa = 4.27TGFSTLYY271 pKa = 9.71GQPGQVTVYY280 pKa = 10.68SQIFDD285 pKa = 3.76WLNSQWDD292 pKa = 4.01TGHH295 pKa = 5.92KK296 pKa = 8.56TVPLFAFDD304 pKa = 6.11AFDD307 pKa = 5.55QPAAWPDD314 pKa = 3.15VEE316 pKa = 4.85KK317 pKa = 11.17YY318 pKa = 10.45YY319 pKa = 11.14GIYY322 pKa = 9.42TDD324 pKa = 4.48SGSLKK329 pKa = 10.14PGLAAILPSWLTTATSTYY347 pKa = 10.83VEE349 pKa = 4.23GTAAAEE355 pKa = 4.06RR356 pKa = 11.84FFAKK360 pKa = 10.08PGNDD364 pKa = 2.76IVDD367 pKa = 4.18PGLGDD372 pKa = 4.38DD373 pKa = 4.05VLALGGTSARR383 pKa = 11.84YY384 pKa = 9.16SVALSGAVALVTDD397 pKa = 4.8LMTNSTDD404 pKa = 3.36VVSNVQALEE413 pKa = 4.02FADD416 pKa = 3.92GTKK419 pKa = 10.87ALFATDD425 pKa = 3.49QTHH428 pKa = 6.49IAALYY433 pKa = 9.33EE434 pKa = 4.22AMLGRR439 pKa = 11.84DD440 pKa = 3.56ADD442 pKa = 4.07SAGLAFWASQSFSGTTFDD460 pKa = 4.83AMASAFTDD468 pKa = 3.44SAEE471 pKa = 4.01YY472 pKa = 10.15AARR475 pKa = 11.84STLDD479 pKa = 3.28DD480 pKa = 4.09STWLAGLYY488 pKa = 10.62DD489 pKa = 4.13NLLHH493 pKa = 7.14RR494 pKa = 11.84AADD497 pKa = 4.06SEE499 pKa = 4.42GQAYY503 pKa = 8.54WLAQRR508 pKa = 11.84DD509 pKa = 3.77AGVARR514 pKa = 11.84HH515 pKa = 5.97EE516 pKa = 4.31IEE518 pKa = 4.03TTFILSAEE526 pKa = 4.29FTSAAHH532 pKa = 6.71DD533 pKa = 4.42LVDD536 pKa = 3.47STQWMGLVGGTLPDD550 pKa = 3.68LPAAHH555 pKa = 6.81GPSSS559 pKa = 3.26
MM1 pKa = 7.44ATTDD5 pKa = 3.63AASSSLIPAHH15 pKa = 6.81SPFSYY20 pKa = 10.73NVGINYY26 pKa = 7.91EE27 pKa = 4.08SWDD30 pKa = 3.51GGRR33 pKa = 11.84VGRR36 pKa = 11.84VIANDD41 pKa = 3.69LDD43 pKa = 5.89AITQNFKK50 pKa = 11.09LIRR53 pKa = 11.84TYY55 pKa = 11.15HH56 pKa = 6.48DD57 pKa = 3.43AAVGTSDD64 pKa = 3.21PTAPIIEE71 pKa = 4.31PTQQEE76 pKa = 4.46VISYY80 pKa = 9.26VLAHH84 pKa = 6.99PGLEE88 pKa = 3.99LSMGTNNNALANGGYY103 pKa = 8.93GAPWSPGLMVDD114 pKa = 5.25PAYY117 pKa = 9.84TDD119 pKa = 2.85QWVAMIIEE127 pKa = 4.75AFGSTNAVEE136 pKa = 5.02EE137 pKa = 4.08NLKK140 pKa = 10.18MILLGNEE147 pKa = 3.83LDD149 pKa = 4.51ANGPPPGDD157 pKa = 3.51PLFEE161 pKa = 4.26TYY163 pKa = 10.76YY164 pKa = 10.85SVWIPTAFQNLQTSLANAGLGSIPISTTIANYY196 pKa = 8.14GTGNVVSTSISSFIEE211 pKa = 4.55TNWSASWNGGEE222 pKa = 5.26PIVLYY227 pKa = 10.51NQYY230 pKa = 9.2TPDD233 pKa = 3.72NGSSTDD239 pKa = 3.51YY240 pKa = 11.1SSVISYY246 pKa = 10.08FEE248 pKa = 4.12SLATTFDD255 pKa = 3.3GALAPFIGEE264 pKa = 4.27TGFSTLYY271 pKa = 9.71GQPGQVTVYY280 pKa = 10.68SQIFDD285 pKa = 3.76WLNSQWDD292 pKa = 4.01TGHH295 pKa = 5.92KK296 pKa = 8.56TVPLFAFDD304 pKa = 6.11AFDD307 pKa = 5.55QPAAWPDD314 pKa = 3.15VEE316 pKa = 4.85KK317 pKa = 11.17YY318 pKa = 10.45YY319 pKa = 11.14GIYY322 pKa = 9.42TDD324 pKa = 4.48SGSLKK329 pKa = 10.14PGLAAILPSWLTTATSTYY347 pKa = 10.83VEE349 pKa = 4.23GTAAAEE355 pKa = 4.06RR356 pKa = 11.84FFAKK360 pKa = 10.08PGNDD364 pKa = 2.76IVDD367 pKa = 4.18PGLGDD372 pKa = 4.38DD373 pKa = 4.05VLALGGTSARR383 pKa = 11.84YY384 pKa = 9.16SVALSGAVALVTDD397 pKa = 4.8LMTNSTDD404 pKa = 3.36VVSNVQALEE413 pKa = 4.02FADD416 pKa = 3.92GTKK419 pKa = 10.87ALFATDD425 pKa = 3.49QTHH428 pKa = 6.49IAALYY433 pKa = 9.33EE434 pKa = 4.22AMLGRR439 pKa = 11.84DD440 pKa = 3.56ADD442 pKa = 4.07SAGLAFWASQSFSGTTFDD460 pKa = 4.83AMASAFTDD468 pKa = 3.44SAEE471 pKa = 4.01YY472 pKa = 10.15AARR475 pKa = 11.84STLDD479 pKa = 3.28DD480 pKa = 4.09STWLAGLYY488 pKa = 10.62DD489 pKa = 4.13NLLHH493 pKa = 7.14RR494 pKa = 11.84AADD497 pKa = 4.06SEE499 pKa = 4.42GQAYY503 pKa = 8.54WLAQRR508 pKa = 11.84DD509 pKa = 3.77AGVARR514 pKa = 11.84HH515 pKa = 5.97EE516 pKa = 4.31IEE518 pKa = 4.03TTFILSAEE526 pKa = 4.29FTSAAHH532 pKa = 6.71DD533 pKa = 4.42LVDD536 pKa = 3.47STQWMGLVGGTLPDD550 pKa = 3.68LPAAHH555 pKa = 6.81GPSSS559 pKa = 3.26
Molecular weight: 59.35 kDa
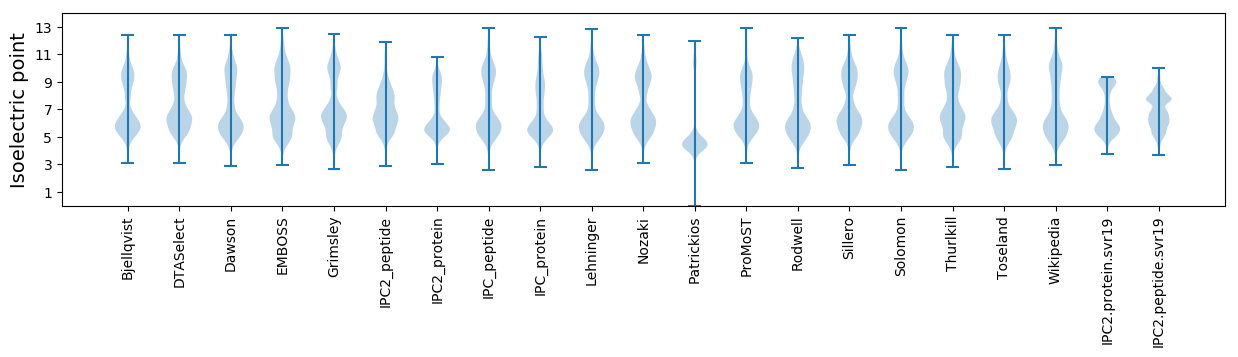
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V3UHC2|A0A2V3UHC2_9RHIZ Elongation factor Tu (Fragment) OS=Chelatococcus asaccharovorans OX=28210 GN=C7450_106425 PE=3 SV=1
MM1 pKa = 7.46ARR3 pKa = 11.84SQAVASGVGQPVSAQPASVLVGTAAFGAPLLVLALGHH40 pKa = 6.14MLSNMLRR47 pKa = 11.84TLPAIATDD55 pKa = 3.61VMAVDD60 pKa = 4.12IGVSAEE66 pKa = 3.95ALASLTGAYY75 pKa = 9.71HH76 pKa = 6.33FAFAAGQIPVGVALDD91 pKa = 3.83RR92 pKa = 11.84YY93 pKa = 9.13GVRR96 pKa = 11.84TVSLTLFAIVTVGAALAAVVGGAAGFFLAQIVLGVGCCGMLLCPMTLAAKK146 pKa = 10.39LLTPAKK152 pKa = 9.78FGLWSGLIQGVGNSGMLLSASPLAWLIEE180 pKa = 4.13MAGWRR185 pKa = 11.84AGYY188 pKa = 9.71GVAAIAGVVIAGLVFLLVPNTPPEE212 pKa = 4.39RR213 pKa = 11.84GAGHH217 pKa = 5.92ATLRR221 pKa = 11.84SEE223 pKa = 3.97ARR225 pKa = 11.84EE226 pKa = 3.9VVQIGLSARR235 pKa = 11.84MRR237 pKa = 11.84GIIILALVSFAVMIGIRR254 pKa = 11.84GLWGGPWLMDD264 pKa = 3.16VKK266 pKa = 10.9GLSRR270 pKa = 11.84VEE272 pKa = 3.96AGAVLMPLTIALVLGPIAFGMLDD295 pKa = 3.02RR296 pKa = 11.84RR297 pKa = 11.84IGHH300 pKa = 6.21RR301 pKa = 11.84RR302 pKa = 11.84ALIIAGHH309 pKa = 5.99AVAGAMLLLLAMGGPAGTLSRR330 pKa = 11.84LLGEE334 pKa = 4.15MSLPVTFDD342 pKa = 3.13TTVLFIFGATIAVQPLLFAMGRR364 pKa = 11.84SIFTPDD370 pKa = 3.11KK371 pKa = 10.77AGKK374 pKa = 9.72ALAAINFAFFAGAAIFQPVTGAVSAVWGTAGVIVFLGLLMLTCTLAFALMTEE426 pKa = 4.39RR427 pKa = 11.84RR428 pKa = 11.84ATGGRR433 pKa = 3.5
MM1 pKa = 7.46ARR3 pKa = 11.84SQAVASGVGQPVSAQPASVLVGTAAFGAPLLVLALGHH40 pKa = 6.14MLSNMLRR47 pKa = 11.84TLPAIATDD55 pKa = 3.61VMAVDD60 pKa = 4.12IGVSAEE66 pKa = 3.95ALASLTGAYY75 pKa = 9.71HH76 pKa = 6.33FAFAAGQIPVGVALDD91 pKa = 3.83RR92 pKa = 11.84YY93 pKa = 9.13GVRR96 pKa = 11.84TVSLTLFAIVTVGAALAAVVGGAAGFFLAQIVLGVGCCGMLLCPMTLAAKK146 pKa = 10.39LLTPAKK152 pKa = 9.78FGLWSGLIQGVGNSGMLLSASPLAWLIEE180 pKa = 4.13MAGWRR185 pKa = 11.84AGYY188 pKa = 9.71GVAAIAGVVIAGLVFLLVPNTPPEE212 pKa = 4.39RR213 pKa = 11.84GAGHH217 pKa = 5.92ATLRR221 pKa = 11.84SEE223 pKa = 3.97ARR225 pKa = 11.84EE226 pKa = 3.9VVQIGLSARR235 pKa = 11.84MRR237 pKa = 11.84GIIILALVSFAVMIGIRR254 pKa = 11.84GLWGGPWLMDD264 pKa = 3.16VKK266 pKa = 10.9GLSRR270 pKa = 11.84VEE272 pKa = 3.96AGAVLMPLTIALVLGPIAFGMLDD295 pKa = 3.02RR296 pKa = 11.84RR297 pKa = 11.84IGHH300 pKa = 6.21RR301 pKa = 11.84RR302 pKa = 11.84ALIIAGHH309 pKa = 5.99AVAGAMLLLLAMGGPAGTLSRR330 pKa = 11.84LLGEE334 pKa = 4.15MSLPVTFDD342 pKa = 3.13TTVLFIFGATIAVQPLLFAMGRR364 pKa = 11.84SIFTPDD370 pKa = 3.11KK371 pKa = 10.77AGKK374 pKa = 9.72ALAAINFAFFAGAAIFQPVTGAVSAVWGTAGVIVFLGLLMLTCTLAFALMTEE426 pKa = 4.39RR427 pKa = 11.84RR428 pKa = 11.84ATGGRR433 pKa = 3.5
Molecular weight: 44.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2107697 |
28 |
2242 |
316.5 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.115 ± 0.042 | 0.807 ± 0.009 |
5.553 ± 0.024 | 5.25 ± 0.026 |
3.739 ± 0.02 | 8.672 ± 0.027 |
2.024 ± 0.014 | 5.347 ± 0.019 |
2.893 ± 0.024 | 10.327 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.012 | 2.407 ± 0.018 |
5.435 ± 0.023 | 2.911 ± 0.015 |
7.366 ± 0.031 | 5.344 ± 0.019 |
5.294 ± 0.017 | 7.652 ± 0.024 |
1.293 ± 0.011 | 2.163 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
