
Oryza rufipogon (Brownbeard rice) (Asian wild rice)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
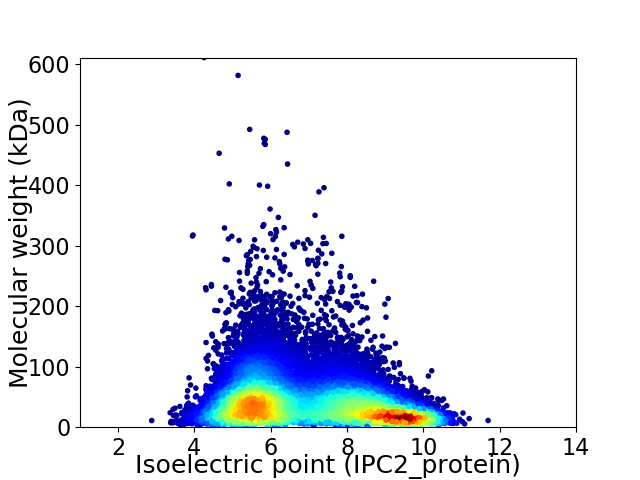
Virtual 2D-PAGE plot for 45598 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E0Q5E9|A0A0E0Q5E9_ORYRU WD_REPEATS_REGION domain-containing protein OS=Oryza rufipogon OX=4529 PE=4 SV=1
MM1 pKa = 7.37CVDD4 pKa = 4.03GEE6 pKa = 4.31RR7 pKa = 11.84LVVPDD12 pKa = 3.89GGGGGGVEE20 pKa = 4.22IAWRR24 pKa = 11.84KK25 pKa = 8.95YY26 pKa = 8.22VLSYY30 pKa = 10.48FADD33 pKa = 4.13GEE35 pKa = 4.45KK36 pKa = 10.86GSSGWVMHH44 pKa = 6.86EE45 pKa = 3.95YY46 pKa = 10.58AITSPADD53 pKa = 3.53LASSAMRR60 pKa = 11.84LYY62 pKa = 10.66RR63 pKa = 11.84IRR65 pKa = 11.84FSGHH69 pKa = 3.62GKK71 pKa = 8.92KK72 pKa = 9.87RR73 pKa = 11.84KK74 pKa = 9.58RR75 pKa = 11.84EE76 pKa = 3.93PDD78 pKa = 3.25SQSAHH83 pKa = 6.89DD84 pKa = 3.42EE85 pKa = 4.12HH86 pKa = 7.96GRR88 pKa = 11.84ARR90 pKa = 11.84CAPQIAMPEE99 pKa = 4.09TALLEE104 pKa = 4.83DD105 pKa = 4.43SAPPPQPVHH114 pKa = 6.54PPAAVVDD121 pKa = 4.53CVCDD125 pKa = 3.63VTDD128 pKa = 3.6QGSSLVFPDD137 pKa = 3.63QPGSIYY143 pKa = 10.52EE144 pKa = 4.67DD145 pKa = 3.72EE146 pKa = 4.64LQSFVPEE153 pKa = 4.15FAARR157 pKa = 11.84NLFVSLRR164 pKa = 11.84EE165 pKa = 3.84GSRR168 pKa = 11.84DD169 pKa = 3.6VVAEE173 pKa = 3.78AALIEE178 pKa = 4.61DD179 pKa = 5.02LALSPQPVPPPAEE192 pKa = 4.15VVNQADD198 pKa = 4.18DD199 pKa = 4.34SDD201 pKa = 4.81GADD204 pKa = 3.28QGCSSVFAALPDD216 pKa = 4.82LIVLPPEE223 pKa = 4.24EE224 pKa = 4.6ACGSGGAAPAPSWASSLDD242 pKa = 3.62NQNDD246 pKa = 3.55DD247 pKa = 3.62APAFFEE253 pKa = 4.94FPEE256 pKa = 4.57SMDD259 pKa = 4.42DD260 pKa = 3.58MVGCFDD266 pKa = 4.6FASMDD271 pKa = 3.77NQSCTSAVSEE281 pKa = 3.99IAVLEE286 pKa = 4.43EE287 pKa = 4.44PFLPPPTMVNHH298 pKa = 7.24DD299 pKa = 4.31NNSVSDD305 pKa = 4.5GADD308 pKa = 3.03QSCFGVGDD316 pKa = 3.51NSTLVFSDD324 pKa = 3.27LTGSIDD330 pKa = 3.71EE331 pKa = 5.59DD332 pKa = 3.78EE333 pKa = 4.85LQSFVPEE340 pKa = 4.49FVSLPQGSCEE350 pKa = 4.15ADD352 pKa = 3.08AEE354 pKa = 4.36ADD356 pKa = 3.56SGGGVAPAQFAEE368 pKa = 4.51FGGPEE373 pKa = 4.6SMDD376 pKa = 3.77DD377 pKa = 3.73PLNFPAEE384 pKa = 3.99ASGGGDD390 pKa = 2.83RR391 pKa = 11.84AAPASSWVSSQDD403 pKa = 3.43NQNDD407 pKa = 3.58EE408 pKa = 4.34APMFFEE414 pKa = 5.49LPEE417 pKa = 4.43SLDD420 pKa = 3.84DD421 pKa = 3.97MVGCFDD427 pKa = 4.37FAAMDD432 pKa = 4.16GQSCTSAVSEE442 pKa = 4.05TALIEE447 pKa = 4.22EE448 pKa = 4.85LVLPPAAMVNHH459 pKa = 7.04HH460 pKa = 7.66DD461 pKa = 5.13DD462 pKa = 3.97SVSDD466 pKa = 3.83IADD469 pKa = 3.88HH470 pKa = 6.34GCSGAVPPPNSEE482 pKa = 4.57VVDD485 pKa = 4.49LPNDD489 pKa = 3.58SVGADD494 pKa = 3.54QSCSGMVDD502 pKa = 3.65DD503 pKa = 5.66SLSGYY508 pKa = 10.42YY509 pKa = 9.38EE510 pKa = 5.1AEE512 pKa = 4.28LKK514 pKa = 10.54DD515 pKa = 3.68ASGGAGSMMSSPDD528 pKa = 3.13KK529 pKa = 10.82QKK531 pKa = 9.8EE532 pKa = 4.1HH533 pKa = 6.69SSSGVMDD540 pKa = 3.76VEE542 pKa = 4.19ATGFGVPDD550 pKa = 3.59SMDD553 pKa = 3.57GLSCIDD559 pKa = 3.93FAEE562 pKa = 4.32TMDD565 pKa = 5.18DD566 pKa = 4.33LSCIDD571 pKa = 3.63FTIDD575 pKa = 5.64DD576 pKa = 4.56EE577 pKa = 6.63LFDD580 pKa = 3.74LWSS583 pKa = 3.43
MM1 pKa = 7.37CVDD4 pKa = 4.03GEE6 pKa = 4.31RR7 pKa = 11.84LVVPDD12 pKa = 3.89GGGGGGVEE20 pKa = 4.22IAWRR24 pKa = 11.84KK25 pKa = 8.95YY26 pKa = 8.22VLSYY30 pKa = 10.48FADD33 pKa = 4.13GEE35 pKa = 4.45KK36 pKa = 10.86GSSGWVMHH44 pKa = 6.86EE45 pKa = 3.95YY46 pKa = 10.58AITSPADD53 pKa = 3.53LASSAMRR60 pKa = 11.84LYY62 pKa = 10.66RR63 pKa = 11.84IRR65 pKa = 11.84FSGHH69 pKa = 3.62GKK71 pKa = 8.92KK72 pKa = 9.87RR73 pKa = 11.84KK74 pKa = 9.58RR75 pKa = 11.84EE76 pKa = 3.93PDD78 pKa = 3.25SQSAHH83 pKa = 6.89DD84 pKa = 3.42EE85 pKa = 4.12HH86 pKa = 7.96GRR88 pKa = 11.84ARR90 pKa = 11.84CAPQIAMPEE99 pKa = 4.09TALLEE104 pKa = 4.83DD105 pKa = 4.43SAPPPQPVHH114 pKa = 6.54PPAAVVDD121 pKa = 4.53CVCDD125 pKa = 3.63VTDD128 pKa = 3.6QGSSLVFPDD137 pKa = 3.63QPGSIYY143 pKa = 10.52EE144 pKa = 4.67DD145 pKa = 3.72EE146 pKa = 4.64LQSFVPEE153 pKa = 4.15FAARR157 pKa = 11.84NLFVSLRR164 pKa = 11.84EE165 pKa = 3.84GSRR168 pKa = 11.84DD169 pKa = 3.6VVAEE173 pKa = 3.78AALIEE178 pKa = 4.61DD179 pKa = 5.02LALSPQPVPPPAEE192 pKa = 4.15VVNQADD198 pKa = 4.18DD199 pKa = 4.34SDD201 pKa = 4.81GADD204 pKa = 3.28QGCSSVFAALPDD216 pKa = 4.82LIVLPPEE223 pKa = 4.24EE224 pKa = 4.6ACGSGGAAPAPSWASSLDD242 pKa = 3.62NQNDD246 pKa = 3.55DD247 pKa = 3.62APAFFEE253 pKa = 4.94FPEE256 pKa = 4.57SMDD259 pKa = 4.42DD260 pKa = 3.58MVGCFDD266 pKa = 4.6FASMDD271 pKa = 3.77NQSCTSAVSEE281 pKa = 3.99IAVLEE286 pKa = 4.43EE287 pKa = 4.44PFLPPPTMVNHH298 pKa = 7.24DD299 pKa = 4.31NNSVSDD305 pKa = 4.5GADD308 pKa = 3.03QSCFGVGDD316 pKa = 3.51NSTLVFSDD324 pKa = 3.27LTGSIDD330 pKa = 3.71EE331 pKa = 5.59DD332 pKa = 3.78EE333 pKa = 4.85LQSFVPEE340 pKa = 4.49FVSLPQGSCEE350 pKa = 4.15ADD352 pKa = 3.08AEE354 pKa = 4.36ADD356 pKa = 3.56SGGGVAPAQFAEE368 pKa = 4.51FGGPEE373 pKa = 4.6SMDD376 pKa = 3.77DD377 pKa = 3.73PLNFPAEE384 pKa = 3.99ASGGGDD390 pKa = 2.83RR391 pKa = 11.84AAPASSWVSSQDD403 pKa = 3.43NQNDD407 pKa = 3.58EE408 pKa = 4.34APMFFEE414 pKa = 5.49LPEE417 pKa = 4.43SLDD420 pKa = 3.84DD421 pKa = 3.97MVGCFDD427 pKa = 4.37FAAMDD432 pKa = 4.16GQSCTSAVSEE442 pKa = 4.05TALIEE447 pKa = 4.22EE448 pKa = 4.85LVLPPAAMVNHH459 pKa = 7.04HH460 pKa = 7.66DD461 pKa = 5.13DD462 pKa = 3.97SVSDD466 pKa = 3.83IADD469 pKa = 3.88HH470 pKa = 6.34GCSGAVPPPNSEE482 pKa = 4.57VVDD485 pKa = 4.49LPNDD489 pKa = 3.58SVGADD494 pKa = 3.54QSCSGMVDD502 pKa = 3.65DD503 pKa = 5.66SLSGYY508 pKa = 10.42YY509 pKa = 9.38EE510 pKa = 5.1AEE512 pKa = 4.28LKK514 pKa = 10.54DD515 pKa = 3.68ASGGAGSMMSSPDD528 pKa = 3.13KK529 pKa = 10.82QKK531 pKa = 9.8EE532 pKa = 4.1HH533 pKa = 6.69SSSGVMDD540 pKa = 3.76VEE542 pKa = 4.19ATGFGVPDD550 pKa = 3.59SMDD553 pKa = 3.57GLSCIDD559 pKa = 3.93FAEE562 pKa = 4.32TMDD565 pKa = 5.18DD566 pKa = 4.33LSCIDD571 pKa = 3.63FTIDD575 pKa = 5.64DD576 pKa = 4.56EE577 pKa = 6.63LFDD580 pKa = 3.74LWSS583 pKa = 3.43
Molecular weight: 61.2 kDa
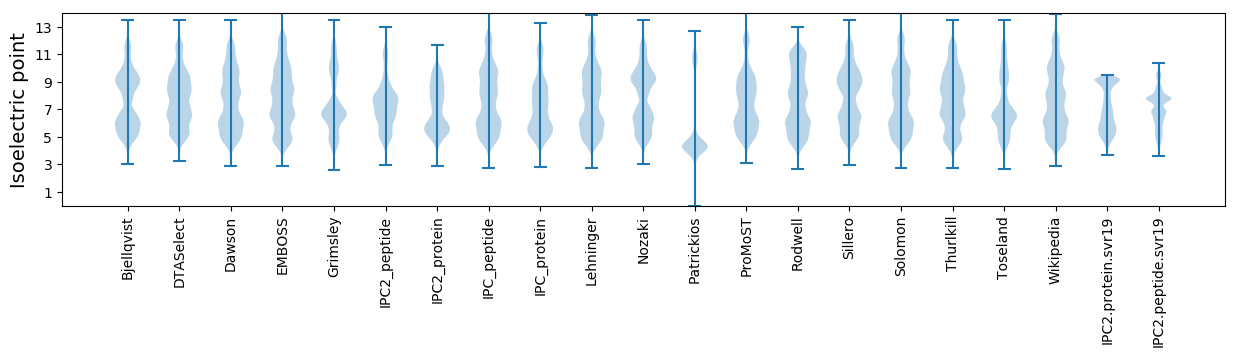
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E0QWC7|A0A0E0QWC7_ORYRU Protein kinase domain-containing protein OS=Oryza rufipogon OX=4529 PE=3 SV=1
MM1 pKa = 7.86DD2 pKa = 5.3KK3 pKa = 10.66KK4 pKa = 10.34HH5 pKa = 6.03STACLHH11 pKa = 6.56RR12 pKa = 11.84PTTGRR17 pKa = 11.84RR18 pKa = 11.84GQAGRR23 pKa = 11.84RR24 pKa = 11.84GRR26 pKa = 11.84AAGRR30 pKa = 11.84RR31 pKa = 11.84SALWRR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84TSSPPRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PPSPPPPPPRR58 pKa = 11.84SAPAPWRR65 pKa = 11.84TRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84TAAAAAPPPRR80 pKa = 11.84TRR82 pKa = 11.84TPPRR86 pKa = 11.84PPPAAAGTAGGSRR99 pKa = 11.84SPSGRR104 pKa = 11.84RR105 pKa = 11.84TAAAPRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84TPRR116 pKa = 11.84GTPPPASSTPAAGTGPASSS135 pKa = 3.73
MM1 pKa = 7.86DD2 pKa = 5.3KK3 pKa = 10.66KK4 pKa = 10.34HH5 pKa = 6.03STACLHH11 pKa = 6.56RR12 pKa = 11.84PTTGRR17 pKa = 11.84RR18 pKa = 11.84GQAGRR23 pKa = 11.84RR24 pKa = 11.84GRR26 pKa = 11.84AAGRR30 pKa = 11.84RR31 pKa = 11.84SALWRR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84TSSPPRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PPSPPPPPPRR58 pKa = 11.84SAPAPWRR65 pKa = 11.84TRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84TAAAAAPPPRR80 pKa = 11.84TRR82 pKa = 11.84TPPRR86 pKa = 11.84PPPAAAGTAGGSRR99 pKa = 11.84SPSGRR104 pKa = 11.84RR105 pKa = 11.84TAAAPRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84TPRR116 pKa = 11.84GTPPPASSTPAAGTGPASSS135 pKa = 3.73
Molecular weight: 14.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
18680246 |
7 |
6304 |
409.7 |
45.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.171 ± 0.018 | 1.94 ± 0.006 |
5.388 ± 0.008 | 5.989 ± 0.013 |
3.6 ± 0.007 | 7.43 ± 0.013 |
2.51 ± 0.005 | 4.495 ± 0.01 |
4.836 ± 0.011 | 9.55 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.004 | 3.549 ± 0.009 |
5.542 ± 0.013 | 3.397 ± 0.008 |
6.443 ± 0.013 | 8.347 ± 0.015 |
4.805 ± 0.007 | 6.703 ± 0.009 |
1.328 ± 0.004 | 2.536 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
