
Promicromonospora thailandica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Promicromonospora
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
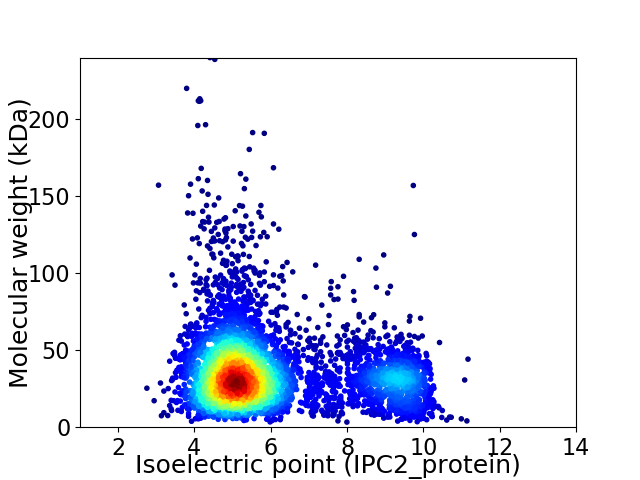
Virtual 2D-PAGE plot for 5167 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
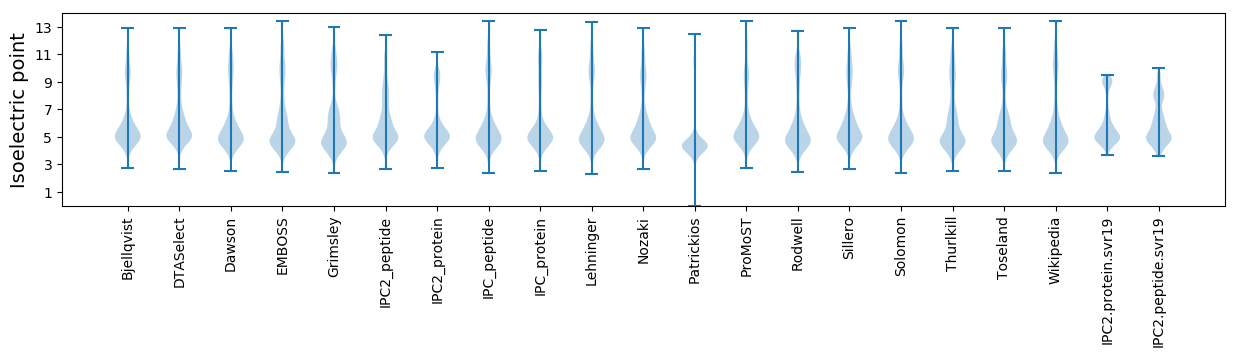
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A397LR42|A0A397LR42_9MICO 8-oxo-dGTP pyrophosphatase MutT (NUDIX family) OS=Promicromonospora thailandica OX=765201 GN=C7338_3333 PE=4 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84QQLSRR7 pKa = 11.84LLAGLAVSALVALPLASTGAVAAEE31 pKa = 3.95PAEE34 pKa = 4.37EE35 pKa = 5.57GVTAWSPPAKK45 pKa = 10.0SAPKK49 pKa = 10.17DD50 pKa = 3.58RR51 pKa = 11.84APARR55 pKa = 11.84ADD57 pKa = 3.39GATTAAAEE65 pKa = 4.32VADD68 pKa = 5.49QVLTWTADD76 pKa = 3.4GGVSEE81 pKa = 4.82YY82 pKa = 11.39ASAPATATAGATTIVFEE99 pKa = 4.26NSRR102 pKa = 11.84ATGNNIAMSHH112 pKa = 5.77TLTFDD117 pKa = 3.08TSTPGYY123 pKa = 9.43NHH125 pKa = 7.54DD126 pKa = 3.71VTLDD130 pKa = 3.35ILANPLDD137 pKa = 4.49ANGGRR142 pKa = 11.84YY143 pKa = 7.6EE144 pKa = 4.31AEE146 pKa = 4.1VVLTPGTYY154 pKa = 9.31RR155 pKa = 11.84YY156 pKa = 9.34FCAIPGHH163 pKa = 4.92STMVGEE169 pKa = 4.47LVVTEE174 pKa = 4.69GPGEE178 pKa = 4.2DD179 pKa = 3.06TTAPVVTPQVAGQQDD194 pKa = 3.57GTGAYY199 pKa = 9.17IGTATVSLAAEE210 pKa = 4.38DD211 pKa = 5.25DD212 pKa = 3.65ISGVASVEE220 pKa = 4.0YY221 pKa = 10.55ALDD224 pKa = 3.83GGEE227 pKa = 3.88FAPYY231 pKa = 10.23GEE233 pKa = 4.39PVVLDD238 pKa = 3.73EE239 pKa = 5.17PGTYY243 pKa = 10.43SLAYY247 pKa = 9.98RR248 pKa = 11.84ATDD251 pKa = 3.16NAGNVSEE258 pKa = 5.23PGTLDD263 pKa = 3.34VTVVSADD270 pKa = 3.93GEE272 pKa = 4.52DD273 pKa = 3.52TTAPTVTAEE282 pKa = 4.19VTGEE286 pKa = 4.0LDD288 pKa = 4.18DD289 pKa = 5.33DD290 pKa = 4.2GAYY293 pKa = 10.69VGTATVNLAALDD305 pKa = 4.15AGSGVASVEE314 pKa = 3.8YY315 pKa = 10.71DD316 pKa = 3.3LDD318 pKa = 3.98GAGYY322 pKa = 10.01AAYY325 pKa = 8.39TDD327 pKa = 4.24PVAVDD332 pKa = 3.43TPGEE336 pKa = 4.07HH337 pKa = 5.37TVTYY341 pKa = 10.37RR342 pKa = 11.84ATDD345 pKa = 3.24NAGNVSEE352 pKa = 4.97PGSATFTVVEE362 pKa = 4.71PSQEE366 pKa = 4.08DD367 pKa = 3.84TTPPTVTAHH376 pKa = 6.51VMGEE380 pKa = 3.9QDD382 pKa = 2.94GDD384 pKa = 3.65GAYY387 pKa = 10.32VGSATVHH394 pKa = 6.28LVAEE398 pKa = 4.79DD399 pKa = 3.59EE400 pKa = 4.4ASEE403 pKa = 4.41VTSVEE408 pKa = 3.78YY409 pKa = 10.97DD410 pKa = 3.16LDD412 pKa = 4.05GAGWTDD418 pKa = 3.23YY419 pKa = 11.28GDD421 pKa = 3.83PVLVDD426 pKa = 3.03VTGSHH431 pKa = 6.04TLTYY435 pKa = 10.35RR436 pKa = 11.84ATDD439 pKa = 3.28AAGNTSEE446 pKa = 4.59PGSVEE451 pKa = 3.66IVVVGGADD459 pKa = 3.71PPDD462 pKa = 3.93TTPPIASAQVDD473 pKa = 4.09GDD475 pKa = 3.48QDD477 pKa = 3.85DD478 pKa = 4.06YY479 pKa = 12.07GVYY482 pKa = 10.71LEE484 pKa = 4.52VATVSISARR493 pKa = 11.84DD494 pKa = 3.91DD495 pKa = 3.52EE496 pKa = 5.08SGVAFVEE503 pKa = 4.35WALGDD508 pKa = 3.66TAFRR512 pKa = 11.84EE513 pKa = 4.27YY514 pKa = 10.4TGPFDD519 pKa = 3.65VTDD522 pKa = 4.84PGAHH526 pKa = 6.68ALTYY530 pKa = 10.38RR531 pKa = 11.84ATDD534 pKa = 3.13NAGNVTRR541 pKa = 11.84TGTLRR546 pKa = 11.84FSVSRR551 pKa = 11.84STIDD555 pKa = 3.01ACPVSDD561 pKa = 4.4EE562 pKa = 4.48RR563 pKa = 11.84PTVILGNIDD572 pKa = 3.84SKK574 pKa = 9.75VTNYY578 pKa = 10.95DD579 pKa = 3.24SGNGCTVADD588 pKa = 5.69LIDD591 pKa = 5.16ADD593 pKa = 4.42GAWVSHH599 pKa = 5.66NAFVLHH605 pKa = 6.02VNQVADD611 pKa = 4.27RR612 pKa = 11.84LDD614 pKa = 3.92DD615 pKa = 4.55ADD617 pKa = 4.59VITATEE623 pKa = 3.9ATRR626 pKa = 11.84LIRR629 pKa = 11.84AARR632 pKa = 11.84QSDD635 pKa = 3.71VGRR638 pKa = 4.31
MM1 pKa = 7.48RR2 pKa = 11.84QQLSRR7 pKa = 11.84LLAGLAVSALVALPLASTGAVAAEE31 pKa = 3.95PAEE34 pKa = 4.37EE35 pKa = 5.57GVTAWSPPAKK45 pKa = 10.0SAPKK49 pKa = 10.17DD50 pKa = 3.58RR51 pKa = 11.84APARR55 pKa = 11.84ADD57 pKa = 3.39GATTAAAEE65 pKa = 4.32VADD68 pKa = 5.49QVLTWTADD76 pKa = 3.4GGVSEE81 pKa = 4.82YY82 pKa = 11.39ASAPATATAGATTIVFEE99 pKa = 4.26NSRR102 pKa = 11.84ATGNNIAMSHH112 pKa = 5.77TLTFDD117 pKa = 3.08TSTPGYY123 pKa = 9.43NHH125 pKa = 7.54DD126 pKa = 3.71VTLDD130 pKa = 3.35ILANPLDD137 pKa = 4.49ANGGRR142 pKa = 11.84YY143 pKa = 7.6EE144 pKa = 4.31AEE146 pKa = 4.1VVLTPGTYY154 pKa = 9.31RR155 pKa = 11.84YY156 pKa = 9.34FCAIPGHH163 pKa = 4.92STMVGEE169 pKa = 4.47LVVTEE174 pKa = 4.69GPGEE178 pKa = 4.2DD179 pKa = 3.06TTAPVVTPQVAGQQDD194 pKa = 3.57GTGAYY199 pKa = 9.17IGTATVSLAAEE210 pKa = 4.38DD211 pKa = 5.25DD212 pKa = 3.65ISGVASVEE220 pKa = 4.0YY221 pKa = 10.55ALDD224 pKa = 3.83GGEE227 pKa = 3.88FAPYY231 pKa = 10.23GEE233 pKa = 4.39PVVLDD238 pKa = 3.73EE239 pKa = 5.17PGTYY243 pKa = 10.43SLAYY247 pKa = 9.98RR248 pKa = 11.84ATDD251 pKa = 3.16NAGNVSEE258 pKa = 5.23PGTLDD263 pKa = 3.34VTVVSADD270 pKa = 3.93GEE272 pKa = 4.52DD273 pKa = 3.52TTAPTVTAEE282 pKa = 4.19VTGEE286 pKa = 4.0LDD288 pKa = 4.18DD289 pKa = 5.33DD290 pKa = 4.2GAYY293 pKa = 10.69VGTATVNLAALDD305 pKa = 4.15AGSGVASVEE314 pKa = 3.8YY315 pKa = 10.71DD316 pKa = 3.3LDD318 pKa = 3.98GAGYY322 pKa = 10.01AAYY325 pKa = 8.39TDD327 pKa = 4.24PVAVDD332 pKa = 3.43TPGEE336 pKa = 4.07HH337 pKa = 5.37TVTYY341 pKa = 10.37RR342 pKa = 11.84ATDD345 pKa = 3.24NAGNVSEE352 pKa = 4.97PGSATFTVVEE362 pKa = 4.71PSQEE366 pKa = 4.08DD367 pKa = 3.84TTPPTVTAHH376 pKa = 6.51VMGEE380 pKa = 3.9QDD382 pKa = 2.94GDD384 pKa = 3.65GAYY387 pKa = 10.32VGSATVHH394 pKa = 6.28LVAEE398 pKa = 4.79DD399 pKa = 3.59EE400 pKa = 4.4ASEE403 pKa = 4.41VTSVEE408 pKa = 3.78YY409 pKa = 10.97DD410 pKa = 3.16LDD412 pKa = 4.05GAGWTDD418 pKa = 3.23YY419 pKa = 11.28GDD421 pKa = 3.83PVLVDD426 pKa = 3.03VTGSHH431 pKa = 6.04TLTYY435 pKa = 10.35RR436 pKa = 11.84ATDD439 pKa = 3.28AAGNTSEE446 pKa = 4.59PGSVEE451 pKa = 3.66IVVVGGADD459 pKa = 3.71PPDD462 pKa = 3.93TTPPIASAQVDD473 pKa = 4.09GDD475 pKa = 3.48QDD477 pKa = 3.85DD478 pKa = 4.06YY479 pKa = 12.07GVYY482 pKa = 10.71LEE484 pKa = 4.52VATVSISARR493 pKa = 11.84DD494 pKa = 3.91DD495 pKa = 3.52EE496 pKa = 5.08SGVAFVEE503 pKa = 4.35WALGDD508 pKa = 3.66TAFRR512 pKa = 11.84EE513 pKa = 4.27YY514 pKa = 10.4TGPFDD519 pKa = 3.65VTDD522 pKa = 4.84PGAHH526 pKa = 6.68ALTYY530 pKa = 10.38RR531 pKa = 11.84ATDD534 pKa = 3.13NAGNVTRR541 pKa = 11.84TGTLRR546 pKa = 11.84FSVSRR551 pKa = 11.84STIDD555 pKa = 3.01ACPVSDD561 pKa = 4.4EE562 pKa = 4.48RR563 pKa = 11.84PTVILGNIDD572 pKa = 3.84SKK574 pKa = 9.75VTNYY578 pKa = 10.95DD579 pKa = 3.24SGNGCTVADD588 pKa = 5.69LIDD591 pKa = 5.16ADD593 pKa = 4.42GAWVSHH599 pKa = 5.66NAFVLHH605 pKa = 6.02VNQVADD611 pKa = 4.27RR612 pKa = 11.84LDD614 pKa = 3.92DD615 pKa = 4.55ADD617 pKa = 4.59VITATEE623 pKa = 3.9ATRR626 pKa = 11.84LIRR629 pKa = 11.84AARR632 pKa = 11.84QSDD635 pKa = 3.71VGRR638 pKa = 4.31
Molecular weight: 65.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A397LHD0|A0A397LHD0_9MICO Uncharacterized protein OS=Promicromonospora thailandica OX=765201 GN=C7338_0228 PE=4 SV=1
MM1 pKa = 6.92GTGAAGTTALAAEE14 pKa = 4.45RR15 pKa = 11.84DD16 pKa = 3.97GTTTTVRR23 pKa = 11.84AGPSTGTTVRR33 pKa = 11.84AGTTGPAGTTGLVATTGPAGRR54 pKa = 11.84VAGVSGRR61 pKa = 11.84SGVVTTAGVRR71 pKa = 11.84TVRR74 pKa = 11.84AVRR77 pKa = 11.84TASGATAARR86 pKa = 11.84VTEE89 pKa = 4.05ATAARR94 pKa = 11.84GATGRR99 pKa = 11.84SGGTTATGRR108 pKa = 11.84AGRR111 pKa = 11.84TRR113 pKa = 11.84TGTAAVRR120 pKa = 11.84GVRR123 pKa = 11.84RR124 pKa = 11.84GTTTTGRR131 pKa = 11.84VGRR134 pKa = 11.84SVTGMTGRR142 pKa = 11.84VGRR145 pKa = 11.84SRR147 pKa = 11.84SVAGTVRR154 pKa = 11.84GVTTGWVTGRR164 pKa = 11.84SVVVTTRR171 pKa = 11.84PGARR175 pKa = 11.84TVTAAIVRR183 pKa = 11.84GGPSTVMTGRR193 pKa = 11.84AGPSRR198 pKa = 11.84TVAGIGRR205 pKa = 11.84GVTTGWVTGRR215 pKa = 11.84SGGAVTTVPAVRR227 pKa = 11.84SVTATTGRR235 pKa = 11.84AGRR238 pKa = 11.84SRR240 pKa = 11.84TAVGIGRR247 pKa = 11.84AATTAGARR255 pKa = 11.84SGGVVRR261 pKa = 11.84TGRR264 pKa = 11.84GVRR267 pKa = 11.84STTVATGRR275 pKa = 11.84AARR278 pKa = 11.84TGRR281 pKa = 11.84TAPGVPSVIGVTVRR295 pKa = 11.84AGRR298 pKa = 11.84SRR300 pKa = 11.84SVAGIGRR307 pKa = 11.84GVMTGWVTGRR317 pKa = 11.84SVVVATRR324 pKa = 11.84RR325 pKa = 11.84GVRR328 pKa = 11.84TVTAAIVRR336 pKa = 11.84GGPSTVMTGRR346 pKa = 11.84AGRR349 pKa = 11.84SRR351 pKa = 11.84SVAGIGRR358 pKa = 11.84GVMTAGARR366 pKa = 11.84SEE368 pKa = 4.47GAVTTVPAVRR378 pKa = 11.84SRR380 pKa = 11.84TVATGCAATTVGFRR394 pKa = 11.84SGGTTAVATGASSAGATVLPATATSVGGTSVAGTSGRR431 pKa = 11.84RR432 pKa = 11.84ASRR435 pKa = 11.84NRR437 pKa = 11.84RR438 pKa = 11.84SPRR441 pKa = 11.84TPRR444 pKa = 11.84SVSSTGRR451 pKa = 11.84HH452 pKa = 5.18GLACGAA458 pKa = 4.4
MM1 pKa = 6.92GTGAAGTTALAAEE14 pKa = 4.45RR15 pKa = 11.84DD16 pKa = 3.97GTTTTVRR23 pKa = 11.84AGPSTGTTVRR33 pKa = 11.84AGTTGPAGTTGLVATTGPAGRR54 pKa = 11.84VAGVSGRR61 pKa = 11.84SGVVTTAGVRR71 pKa = 11.84TVRR74 pKa = 11.84AVRR77 pKa = 11.84TASGATAARR86 pKa = 11.84VTEE89 pKa = 4.05ATAARR94 pKa = 11.84GATGRR99 pKa = 11.84SGGTTATGRR108 pKa = 11.84AGRR111 pKa = 11.84TRR113 pKa = 11.84TGTAAVRR120 pKa = 11.84GVRR123 pKa = 11.84RR124 pKa = 11.84GTTTTGRR131 pKa = 11.84VGRR134 pKa = 11.84SVTGMTGRR142 pKa = 11.84VGRR145 pKa = 11.84SRR147 pKa = 11.84SVAGTVRR154 pKa = 11.84GVTTGWVTGRR164 pKa = 11.84SVVVTTRR171 pKa = 11.84PGARR175 pKa = 11.84TVTAAIVRR183 pKa = 11.84GGPSTVMTGRR193 pKa = 11.84AGPSRR198 pKa = 11.84TVAGIGRR205 pKa = 11.84GVTTGWVTGRR215 pKa = 11.84SGGAVTTVPAVRR227 pKa = 11.84SVTATTGRR235 pKa = 11.84AGRR238 pKa = 11.84SRR240 pKa = 11.84TAVGIGRR247 pKa = 11.84AATTAGARR255 pKa = 11.84SGGVVRR261 pKa = 11.84TGRR264 pKa = 11.84GVRR267 pKa = 11.84STTVATGRR275 pKa = 11.84AARR278 pKa = 11.84TGRR281 pKa = 11.84TAPGVPSVIGVTVRR295 pKa = 11.84AGRR298 pKa = 11.84SRR300 pKa = 11.84SVAGIGRR307 pKa = 11.84GVMTGWVTGRR317 pKa = 11.84SVVVATRR324 pKa = 11.84RR325 pKa = 11.84GVRR328 pKa = 11.84TVTAAIVRR336 pKa = 11.84GGPSTVMTGRR346 pKa = 11.84AGRR349 pKa = 11.84SRR351 pKa = 11.84SVAGIGRR358 pKa = 11.84GVMTAGARR366 pKa = 11.84SEE368 pKa = 4.47GAVTTVPAVRR378 pKa = 11.84SRR380 pKa = 11.84TVATGCAATTVGFRR394 pKa = 11.84SGGTTAVATGASSAGATVLPATATSVGGTSVAGTSGRR431 pKa = 11.84RR432 pKa = 11.84ASRR435 pKa = 11.84NRR437 pKa = 11.84RR438 pKa = 11.84SPRR441 pKa = 11.84TPRR444 pKa = 11.84SVSSTGRR451 pKa = 11.84HH452 pKa = 5.18GLACGAA458 pKa = 4.4
Molecular weight: 44.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1812030 |
30 |
2269 |
350.7 |
37.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.331 ± 0.052 | 0.519 ± 0.008 |
6.461 ± 0.031 | 5.475 ± 0.028 |
2.631 ± 0.018 | 9.799 ± 0.034 |
2.043 ± 0.016 | 2.932 ± 0.021 |
1.448 ± 0.021 | 10.066 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.585 ± 0.013 | 1.623 ± 0.017 |
6.135 ± 0.027 | 2.619 ± 0.018 |
7.671 ± 0.04 | 4.93 ± 0.022 |
6.473 ± 0.032 | 9.69 ± 0.037 |
1.609 ± 0.017 | 1.959 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
