
Porcine circovirus 1 (PCV1)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 7.86
Get precalculated fractions of proteins
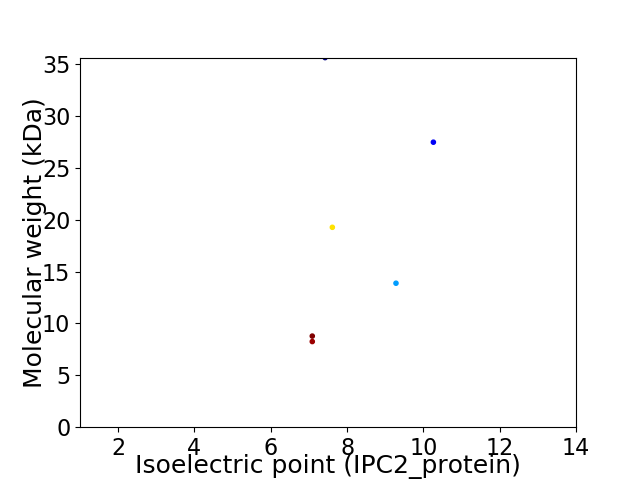
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q80QL5|CAPSD_PCV1 Capsid protein OS=Porcine circovirus 1 OX=133704 GN=Cap PE=3 SV=2
MM1 pKa = 7.76PSKK4 pKa = 10.7KK5 pKa = 10.15SGPQPHH11 pKa = 6.44KK12 pKa = 10.3RR13 pKa = 11.84WVFTLNNPSEE23 pKa = 4.15EE24 pKa = 4.23EE25 pKa = 3.89KK26 pKa = 11.29NKK28 pKa = 9.72IRR30 pKa = 11.84EE31 pKa = 4.19LPISLFDD38 pKa = 3.63YY39 pKa = 8.0FVCGEE44 pKa = 4.09EE45 pKa = 4.15GLEE48 pKa = 4.14EE49 pKa = 4.45GRR51 pKa = 11.84TPHH54 pKa = 6.17LQGFANFAKK63 pKa = 10.18KK64 pKa = 7.57QTFNKK69 pKa = 8.67VKK71 pKa = 8.5WYY73 pKa = 10.26FGARR77 pKa = 11.84CHH79 pKa = 6.85IEE81 pKa = 3.87KK82 pKa = 10.86AKK84 pKa = 9.75GTDD87 pKa = 3.47QQNKK91 pKa = 8.81EE92 pKa = 4.08YY93 pKa = 10.19CSKK96 pKa = 10.39EE97 pKa = 3.48GHH99 pKa = 6.47ILIEE103 pKa = 4.56CGAPRR108 pKa = 11.84NQGKK112 pKa = 10.22RR113 pKa = 11.84SDD115 pKa = 3.69LSTAVSTLLEE125 pKa = 4.4TGSLVTVAEE134 pKa = 4.41QFPVTYY140 pKa = 10.26VRR142 pKa = 11.84NFRR145 pKa = 11.84GLAEE149 pKa = 4.1LLKK152 pKa = 11.05VSGKK156 pKa = 7.49MQQRR160 pKa = 11.84DD161 pKa = 3.45WKK163 pKa = 8.27TAVHH167 pKa = 6.3VIVGPPGCGKK177 pKa = 9.37SQWARR182 pKa = 11.84NFAEE186 pKa = 5.03PSDD189 pKa = 4.45TYY191 pKa = 10.64WKK193 pKa = 10.05PSRR196 pKa = 11.84NKK198 pKa = 8.96WWDD201 pKa = 3.68GYY203 pKa = 10.6HH204 pKa = 6.06GEE206 pKa = 4.32EE207 pKa = 5.11VVVLDD212 pKa = 5.98DD213 pKa = 4.52FYY215 pKa = 11.98GWLPWDD221 pKa = 4.33DD222 pKa = 4.85LLRR225 pKa = 11.84LCDD228 pKa = 4.28RR229 pKa = 11.84YY230 pKa = 10.51PLTVEE235 pKa = 4.37TKK237 pKa = 9.91GGTVPFLARR246 pKa = 11.84SILITSNQAPQEE258 pKa = 4.44WYY260 pKa = 10.74SSTAVPAVEE269 pKa = 3.43ALYY272 pKa = 10.72RR273 pKa = 11.84RR274 pKa = 11.84ITTLQFWKK282 pKa = 9.09TAGEE286 pKa = 4.08QSTEE290 pKa = 3.96VPEE293 pKa = 4.57GRR295 pKa = 11.84FEE297 pKa = 5.48AVDD300 pKa = 4.0PPCALFPYY308 pKa = 9.27KK309 pKa = 10.19INYY312 pKa = 8.38
MM1 pKa = 7.76PSKK4 pKa = 10.7KK5 pKa = 10.15SGPQPHH11 pKa = 6.44KK12 pKa = 10.3RR13 pKa = 11.84WVFTLNNPSEE23 pKa = 4.15EE24 pKa = 4.23EE25 pKa = 3.89KK26 pKa = 11.29NKK28 pKa = 9.72IRR30 pKa = 11.84EE31 pKa = 4.19LPISLFDD38 pKa = 3.63YY39 pKa = 8.0FVCGEE44 pKa = 4.09EE45 pKa = 4.15GLEE48 pKa = 4.14EE49 pKa = 4.45GRR51 pKa = 11.84TPHH54 pKa = 6.17LQGFANFAKK63 pKa = 10.18KK64 pKa = 7.57QTFNKK69 pKa = 8.67VKK71 pKa = 8.5WYY73 pKa = 10.26FGARR77 pKa = 11.84CHH79 pKa = 6.85IEE81 pKa = 3.87KK82 pKa = 10.86AKK84 pKa = 9.75GTDD87 pKa = 3.47QQNKK91 pKa = 8.81EE92 pKa = 4.08YY93 pKa = 10.19CSKK96 pKa = 10.39EE97 pKa = 3.48GHH99 pKa = 6.47ILIEE103 pKa = 4.56CGAPRR108 pKa = 11.84NQGKK112 pKa = 10.22RR113 pKa = 11.84SDD115 pKa = 3.69LSTAVSTLLEE125 pKa = 4.4TGSLVTVAEE134 pKa = 4.41QFPVTYY140 pKa = 10.26VRR142 pKa = 11.84NFRR145 pKa = 11.84GLAEE149 pKa = 4.1LLKK152 pKa = 11.05VSGKK156 pKa = 7.49MQQRR160 pKa = 11.84DD161 pKa = 3.45WKK163 pKa = 8.27TAVHH167 pKa = 6.3VIVGPPGCGKK177 pKa = 9.37SQWARR182 pKa = 11.84NFAEE186 pKa = 5.03PSDD189 pKa = 4.45TYY191 pKa = 10.64WKK193 pKa = 10.05PSRR196 pKa = 11.84NKK198 pKa = 8.96WWDD201 pKa = 3.68GYY203 pKa = 10.6HH204 pKa = 6.06GEE206 pKa = 4.32EE207 pKa = 5.11VVVLDD212 pKa = 5.98DD213 pKa = 4.52FYY215 pKa = 11.98GWLPWDD221 pKa = 4.33DD222 pKa = 4.85LLRR225 pKa = 11.84LCDD228 pKa = 4.28RR229 pKa = 11.84YY230 pKa = 10.51PLTVEE235 pKa = 4.37TKK237 pKa = 9.91GGTVPFLARR246 pKa = 11.84SILITSNQAPQEE258 pKa = 4.44WYY260 pKa = 10.74SSTAVPAVEE269 pKa = 3.43ALYY272 pKa = 10.72RR273 pKa = 11.84RR274 pKa = 11.84ITTLQFWKK282 pKa = 9.09TAGEE286 pKa = 4.08QSTEE290 pKa = 3.96VPEE293 pKa = 4.57GRR295 pKa = 11.84FEE297 pKa = 5.48AVDD300 pKa = 4.0PPCALFPYY308 pKa = 9.27KK309 pKa = 10.19INYY312 pKa = 8.38
Molecular weight: 35.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q805H4-2|REP-2_PCV1 Isoform of Q805H4 Isoform Rep' of Replication-associated protein OS=Porcine circovirus 1 OX=133704 GN=Rep PE=4 SV=1
MM1 pKa = 6.52TWPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 8.64RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84PRR16 pKa = 11.84SHH18 pKa = 7.24LGNILRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84PYY28 pKa = 9.6LAHH31 pKa = 6.14PAFRR35 pKa = 11.84NRR37 pKa = 11.84YY38 pKa = 5.9RR39 pKa = 11.84WRR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 7.69TGIFNSRR50 pKa = 11.84LSTEE54 pKa = 3.98LVLTIKK60 pKa = 10.55GGYY63 pKa = 7.57SQPSWNVNYY72 pKa = 10.68LKK74 pKa = 10.9FNIGQFLPPSGGTNPLPLPFQYY96 pKa = 11.0YY97 pKa = 10.16RR98 pKa = 11.84IRR100 pKa = 11.84KK101 pKa = 8.4AKK103 pKa = 9.93YY104 pKa = 7.89EE105 pKa = 4.13FYY107 pKa = 10.29PRR109 pKa = 11.84DD110 pKa = 4.08PITSNEE116 pKa = 4.04RR117 pKa = 11.84GVGSTVVILDD127 pKa = 3.79ANFVTPSTNLAYY139 pKa = 10.19DD140 pKa = 3.7PYY142 pKa = 10.12INYY145 pKa = 10.06SSRR148 pKa = 11.84HH149 pKa = 5.27TIRR152 pKa = 11.84QPFTYY157 pKa = 9.31HH158 pKa = 5.9SRR160 pKa = 11.84YY161 pKa = 7.46FTPKK165 pKa = 9.93PEE167 pKa = 4.53LDD169 pKa = 3.34QTIDD173 pKa = 3.01WFHH176 pKa = 7.17PNNKK180 pKa = 9.16RR181 pKa = 11.84NQLWLHH187 pKa = 6.5LNTHH191 pKa = 6.09TNVEE195 pKa = 4.43HH196 pKa = 6.47TGLGYY201 pKa = 11.1ALQNAATAQNYY212 pKa = 7.12VVRR215 pKa = 11.84LTIYY219 pKa = 8.42VQFRR223 pKa = 11.84EE224 pKa = 5.18FILKK228 pKa = 10.47DD229 pKa = 3.32PP230 pKa = 4.57
MM1 pKa = 6.52TWPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 8.64RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84PRR16 pKa = 11.84SHH18 pKa = 7.24LGNILRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84PYY28 pKa = 9.6LAHH31 pKa = 6.14PAFRR35 pKa = 11.84NRR37 pKa = 11.84YY38 pKa = 5.9RR39 pKa = 11.84WRR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 7.69TGIFNSRR50 pKa = 11.84LSTEE54 pKa = 3.98LVLTIKK60 pKa = 10.55GGYY63 pKa = 7.57SQPSWNVNYY72 pKa = 10.68LKK74 pKa = 10.9FNIGQFLPPSGGTNPLPLPFQYY96 pKa = 11.0YY97 pKa = 10.16RR98 pKa = 11.84IRR100 pKa = 11.84KK101 pKa = 8.4AKK103 pKa = 9.93YY104 pKa = 7.89EE105 pKa = 4.13FYY107 pKa = 10.29PRR109 pKa = 11.84DD110 pKa = 4.08PITSNEE116 pKa = 4.04RR117 pKa = 11.84GVGSTVVILDD127 pKa = 3.79ANFVTPSTNLAYY139 pKa = 10.19DD140 pKa = 3.7PYY142 pKa = 10.12INYY145 pKa = 10.06SSRR148 pKa = 11.84HH149 pKa = 5.27TIRR152 pKa = 11.84QPFTYY157 pKa = 9.31HH158 pKa = 5.9SRR160 pKa = 11.84YY161 pKa = 7.46FTPKK165 pKa = 9.93PEE167 pKa = 4.53LDD169 pKa = 3.34QTIDD173 pKa = 3.01WFHH176 pKa = 7.17PNNKK180 pKa = 9.16RR181 pKa = 11.84NQLWLHH187 pKa = 6.5LNTHH191 pKa = 6.09TNVEE195 pKa = 4.43HH196 pKa = 6.47TGLGYY201 pKa = 11.1ALQNAATAQNYY212 pKa = 7.12VVRR215 pKa = 11.84LTIYY219 pKa = 8.42VQFRR223 pKa = 11.84EE224 pKa = 5.18FILKK228 pKa = 10.47DD229 pKa = 3.32PP230 pKa = 4.57
Molecular weight: 27.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
987 |
73 |
312 |
164.5 |
18.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.282 ± 1.017 | 1.925 ± 0.711 |
2.736 ± 0.504 | 6.586 ± 1.073 |
4.559 ± 0.393 | 6.079 ± 0.77 |
2.229 ± 0.428 | 4.255 ± 0.43 |
6.586 ± 0.858 | 7.295 ± 0.539 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.811 ± 0.141 | 4.559 ± 0.904 |
8.105 ± 0.808 | 4.661 ± 0.351 |
6.991 ± 1.362 | 6.788 ± 0.654 |
6.991 ± 0.722 | 5.066 ± 0.743 |
2.533 ± 0.346 | 4.965 ± 0.71 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
