
Rhizobium sp. CF142
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
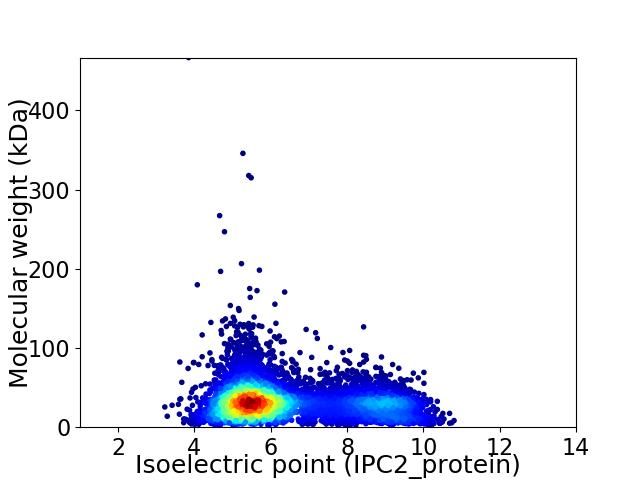
Virtual 2D-PAGE plot for 7229 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J1T8W9|J1T8W9_9RHIZ ABC-type proline/glycine betaine transport system permease component OS=Rhizobium sp. CF142 OX=1144314 GN=PMI11_03582 PE=3 SV=1
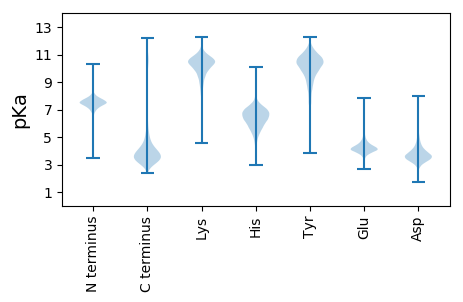
MM1 pKa = 7.71PFVEE5 pKa = 4.45SQVNTFTVNSQYY17 pKa = 10.44STVTTQLPGGGWVITWASTLEE38 pKa = 4.54DD39 pKa = 3.74GSGTGVYY46 pKa = 9.35QQVFDD51 pKa = 4.02ATGSPSGPEE60 pKa = 3.71TRR62 pKa = 11.84VNTIVEE68 pKa = 4.76GNQEE72 pKa = 4.05MPAIAPLADD81 pKa = 4.17GGWVITWGNQGISTPGVYY99 pKa = 9.29QQAFNADD106 pKa = 3.43GTPRR110 pKa = 11.84GSEE113 pKa = 4.17TQVNTYY119 pKa = 9.88EE120 pKa = 4.39GAGQGSRR127 pKa = 11.84SVMGLSDD134 pKa = 4.1GGWVVSWLSVGQDD147 pKa = 3.02GSGYY151 pKa = 10.7GIYY154 pKa = 9.75QQAFAADD161 pKa = 3.89GSKK164 pKa = 10.07IAGEE168 pKa = 4.31TRR170 pKa = 11.84VNTHH174 pKa = 5.53TDD176 pKa = 3.06GNQWLGSMAPLDD188 pKa = 3.94GGGWVATWSSPGQDD202 pKa = 3.14GSEE205 pKa = 3.78TGVFQQIYY213 pKa = 8.93NADD216 pKa = 3.81GTPHH220 pKa = 6.8GGEE223 pKa = 4.24TLVNTYY229 pKa = 9.46TNSYY233 pKa = 9.01QDD235 pKa = 3.47SPSVVARR242 pKa = 11.84NGGWVVIWEE251 pKa = 4.36SLGQDD256 pKa = 3.18GSSRR260 pKa = 11.84GIYY263 pKa = 8.12QQAYY267 pKa = 8.29NADD270 pKa = 3.54GTRR273 pKa = 11.84FGTEE277 pKa = 3.66TRR279 pKa = 11.84VNTATDD285 pKa = 3.42GNEE288 pKa = 4.05AYY290 pKa = 9.93HH291 pKa = 7.11ASAALDD297 pKa = 3.82DD298 pKa = 4.97GGWVVIWISGDD309 pKa = 3.0AHH311 pKa = 7.57LMQQAYY317 pKa = 9.16NANGTPQGGEE327 pKa = 4.14TQVDD331 pKa = 3.4TDD333 pKa = 3.3GTYY336 pKa = 10.54PGNQKK341 pKa = 8.06VTALSDD347 pKa = 3.68GGWVVTWMVIGSSEE361 pKa = 4.21TFYY364 pKa = 11.07HH365 pKa = 7.19VYY367 pKa = 9.69QQAFNADD374 pKa = 3.27GTKK377 pKa = 10.73NGDD380 pKa = 3.27EE381 pKa = 4.11ALINTLTHH389 pKa = 6.31TYY391 pKa = 10.94KK392 pKa = 10.89DD393 pKa = 3.62IPQVAALDD401 pKa = 4.03DD402 pKa = 4.71GGWVVSWASDD412 pKa = 2.99SDD414 pKa = 3.94YY415 pKa = 11.33TDD417 pKa = 3.12VQGYY421 pKa = 10.22DD422 pKa = 2.85SGIFQVRR429 pKa = 11.84FDD431 pKa = 3.97TDD433 pKa = 2.64AHH435 pKa = 5.95AVEE438 pKa = 4.94LPSNRR443 pKa = 11.84IYY445 pKa = 10.35GTYY448 pKa = 9.23PDD450 pKa = 4.34KK451 pKa = 11.44NLVGTAGDD459 pKa = 3.74DD460 pKa = 3.88AIYY463 pKa = 10.55ASSGNDD469 pKa = 3.19TLTGLAGNDD478 pKa = 4.04HH479 pKa = 7.23LDD481 pKa = 3.11GGYY484 pKa = 10.66GHH486 pKa = 7.4DD487 pKa = 4.52RR488 pKa = 11.84MTGGLGDD495 pKa = 3.69DD496 pKa = 4.09TYY498 pKa = 11.31IVDD501 pKa = 4.07TSADD505 pKa = 3.56RR506 pKa = 11.84VVEE509 pKa = 3.88LAGQGTDD516 pKa = 3.37TILVQVSYY524 pKa = 11.45SLGANVEE531 pKa = 4.01NLAFVAEE538 pKa = 4.46GNLYY542 pKa = 10.92ASGNALANVITGNSGNNTLGGAAGSDD568 pKa = 3.66TLSGLDD574 pKa = 3.65GNDD577 pKa = 3.61YY578 pKa = 11.24LDD580 pKa = 4.83GGADD584 pKa = 3.48NDD586 pKa = 3.93KK587 pKa = 11.09LYY589 pKa = 11.09GGSGDD594 pKa = 5.24DD595 pKa = 3.62ILAGGTGDD603 pKa = 3.67DD604 pKa = 4.27TMDD607 pKa = 4.51GGDD610 pKa = 4.04GNDD613 pKa = 3.26KK614 pKa = 11.14YY615 pKa = 11.25KK616 pKa = 10.74VDD618 pKa = 3.66SANDD622 pKa = 3.63VVHH625 pKa = 6.79DD626 pKa = 4.2TGDD629 pKa = 5.18GIWDD633 pKa = 3.73WNDD636 pKa = 3.08IIYY639 pKa = 8.55STAEE643 pKa = 4.16TYY645 pKa = 11.12SLAGTGAEE653 pKa = 4.15TLILQAGAVTGIGDD667 pKa = 3.72SGNNQISGNSAANTLVSGGGSDD689 pKa = 3.72GLAGGGGVDD698 pKa = 3.74TFVLAAPSASNFAFINDD715 pKa = 4.02FGADD719 pKa = 3.64DD720 pKa = 4.34FLAFQSSDD728 pKa = 3.56FQGMTTATLDD738 pKa = 3.45FHH740 pKa = 7.08VGKK743 pKa = 8.3TAIGADD749 pKa = 2.99AQFYY753 pKa = 10.67FNTSDD758 pKa = 3.33RR759 pKa = 11.84TLYY762 pKa = 10.16WDD764 pKa = 4.57DD765 pKa = 5.13DD766 pKa = 4.03GTGGDD771 pKa = 3.63GAVRR775 pKa = 11.84VATLNGAYY783 pKa = 9.82VLQSGDD789 pKa = 3.44FLFAA793 pKa = 5.24
MM1 pKa = 7.71PFVEE5 pKa = 4.45SQVNTFTVNSQYY17 pKa = 10.44STVTTQLPGGGWVITWASTLEE38 pKa = 4.54DD39 pKa = 3.74GSGTGVYY46 pKa = 9.35QQVFDD51 pKa = 4.02ATGSPSGPEE60 pKa = 3.71TRR62 pKa = 11.84VNTIVEE68 pKa = 4.76GNQEE72 pKa = 4.05MPAIAPLADD81 pKa = 4.17GGWVITWGNQGISTPGVYY99 pKa = 9.29QQAFNADD106 pKa = 3.43GTPRR110 pKa = 11.84GSEE113 pKa = 4.17TQVNTYY119 pKa = 9.88EE120 pKa = 4.39GAGQGSRR127 pKa = 11.84SVMGLSDD134 pKa = 4.1GGWVVSWLSVGQDD147 pKa = 3.02GSGYY151 pKa = 10.7GIYY154 pKa = 9.75QQAFAADD161 pKa = 3.89GSKK164 pKa = 10.07IAGEE168 pKa = 4.31TRR170 pKa = 11.84VNTHH174 pKa = 5.53TDD176 pKa = 3.06GNQWLGSMAPLDD188 pKa = 3.94GGGWVATWSSPGQDD202 pKa = 3.14GSEE205 pKa = 3.78TGVFQQIYY213 pKa = 8.93NADD216 pKa = 3.81GTPHH220 pKa = 6.8GGEE223 pKa = 4.24TLVNTYY229 pKa = 9.46TNSYY233 pKa = 9.01QDD235 pKa = 3.47SPSVVARR242 pKa = 11.84NGGWVVIWEE251 pKa = 4.36SLGQDD256 pKa = 3.18GSSRR260 pKa = 11.84GIYY263 pKa = 8.12QQAYY267 pKa = 8.29NADD270 pKa = 3.54GTRR273 pKa = 11.84FGTEE277 pKa = 3.66TRR279 pKa = 11.84VNTATDD285 pKa = 3.42GNEE288 pKa = 4.05AYY290 pKa = 9.93HH291 pKa = 7.11ASAALDD297 pKa = 3.82DD298 pKa = 4.97GGWVVIWISGDD309 pKa = 3.0AHH311 pKa = 7.57LMQQAYY317 pKa = 9.16NANGTPQGGEE327 pKa = 4.14TQVDD331 pKa = 3.4TDD333 pKa = 3.3GTYY336 pKa = 10.54PGNQKK341 pKa = 8.06VTALSDD347 pKa = 3.68GGWVVTWMVIGSSEE361 pKa = 4.21TFYY364 pKa = 11.07HH365 pKa = 7.19VYY367 pKa = 9.69QQAFNADD374 pKa = 3.27GTKK377 pKa = 10.73NGDD380 pKa = 3.27EE381 pKa = 4.11ALINTLTHH389 pKa = 6.31TYY391 pKa = 10.94KK392 pKa = 10.89DD393 pKa = 3.62IPQVAALDD401 pKa = 4.03DD402 pKa = 4.71GGWVVSWASDD412 pKa = 2.99SDD414 pKa = 3.94YY415 pKa = 11.33TDD417 pKa = 3.12VQGYY421 pKa = 10.22DD422 pKa = 2.85SGIFQVRR429 pKa = 11.84FDD431 pKa = 3.97TDD433 pKa = 2.64AHH435 pKa = 5.95AVEE438 pKa = 4.94LPSNRR443 pKa = 11.84IYY445 pKa = 10.35GTYY448 pKa = 9.23PDD450 pKa = 4.34KK451 pKa = 11.44NLVGTAGDD459 pKa = 3.74DD460 pKa = 3.88AIYY463 pKa = 10.55ASSGNDD469 pKa = 3.19TLTGLAGNDD478 pKa = 4.04HH479 pKa = 7.23LDD481 pKa = 3.11GGYY484 pKa = 10.66GHH486 pKa = 7.4DD487 pKa = 4.52RR488 pKa = 11.84MTGGLGDD495 pKa = 3.69DD496 pKa = 4.09TYY498 pKa = 11.31IVDD501 pKa = 4.07TSADD505 pKa = 3.56RR506 pKa = 11.84VVEE509 pKa = 3.88LAGQGTDD516 pKa = 3.37TILVQVSYY524 pKa = 11.45SLGANVEE531 pKa = 4.01NLAFVAEE538 pKa = 4.46GNLYY542 pKa = 10.92ASGNALANVITGNSGNNTLGGAAGSDD568 pKa = 3.66TLSGLDD574 pKa = 3.65GNDD577 pKa = 3.61YY578 pKa = 11.24LDD580 pKa = 4.83GGADD584 pKa = 3.48NDD586 pKa = 3.93KK587 pKa = 11.09LYY589 pKa = 11.09GGSGDD594 pKa = 5.24DD595 pKa = 3.62ILAGGTGDD603 pKa = 3.67DD604 pKa = 4.27TMDD607 pKa = 4.51GGDD610 pKa = 4.04GNDD613 pKa = 3.26KK614 pKa = 11.14YY615 pKa = 11.25KK616 pKa = 10.74VDD618 pKa = 3.66SANDD622 pKa = 3.63VVHH625 pKa = 6.79DD626 pKa = 4.2TGDD629 pKa = 5.18GIWDD633 pKa = 3.73WNDD636 pKa = 3.08IIYY639 pKa = 8.55STAEE643 pKa = 4.16TYY645 pKa = 11.12SLAGTGAEE653 pKa = 4.15TLILQAGAVTGIGDD667 pKa = 3.72SGNNQISGNSAANTLVSGGGSDD689 pKa = 3.72GLAGGGGVDD698 pKa = 3.74TFVLAAPSASNFAFINDD715 pKa = 4.02FGADD719 pKa = 3.64DD720 pKa = 4.34FLAFQSSDD728 pKa = 3.56FQGMTTATLDD738 pKa = 3.45FHH740 pKa = 7.08VGKK743 pKa = 8.3TAIGADD749 pKa = 2.99AQFYY753 pKa = 10.67FNTSDD758 pKa = 3.33RR759 pKa = 11.84TLYY762 pKa = 10.16WDD764 pKa = 4.57DD765 pKa = 5.13DD766 pKa = 4.03GTGGDD771 pKa = 3.63GAVRR775 pKa = 11.84VATLNGAYY783 pKa = 9.82VLQSGDD789 pKa = 3.44FLFAA793 pKa = 5.24
Molecular weight: 82.3 kDa
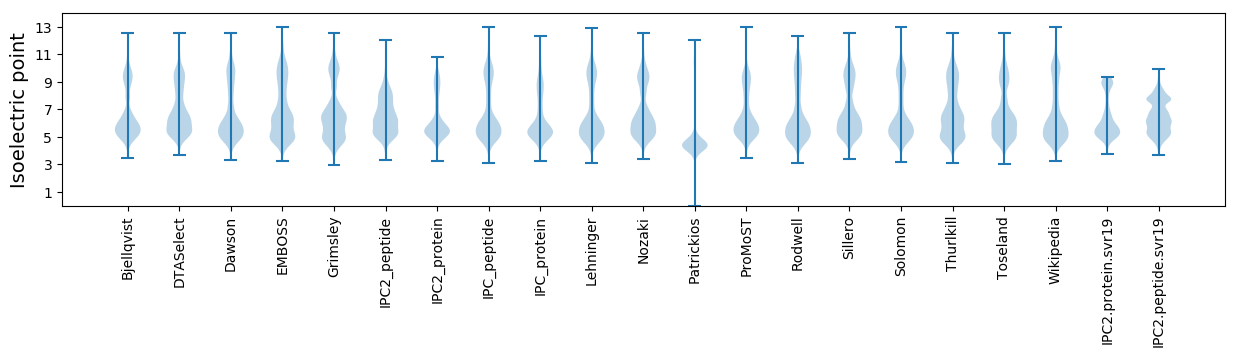
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J2AQL1|J2AQL1_9RHIZ Bifunctional glutamine synthetase adenylyltransferase/adenylyl-removing enzyme OS=Rhizobium sp. CF142 OX=1144314 GN=glnE PE=3 SV=1
MM1 pKa = 7.56ANLVNEE7 pKa = 4.44LVSGVVNSVLKK18 pKa = 10.69EE19 pKa = 3.87ILKK22 pKa = 9.04KK23 pKa = 7.5TTGRR27 pKa = 11.84TTTKK31 pKa = 9.97RR32 pKa = 11.84KK33 pKa = 9.1RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.0AASATTSRR44 pKa = 11.84TTRR47 pKa = 11.84KK48 pKa = 8.21ATPTSSKK55 pKa = 9.87PARR58 pKa = 11.84KK59 pKa = 8.82QVSKK63 pKa = 10.81RR64 pKa = 11.84RR65 pKa = 11.84TAAGRR70 pKa = 11.84SRR72 pKa = 11.84QRR74 pKa = 11.84RR75 pKa = 11.84AA76 pKa = 2.78
MM1 pKa = 7.56ANLVNEE7 pKa = 4.44LVSGVVNSVLKK18 pKa = 10.69EE19 pKa = 3.87ILKK22 pKa = 9.04KK23 pKa = 7.5TTGRR27 pKa = 11.84TTTKK31 pKa = 9.97RR32 pKa = 11.84KK33 pKa = 9.1RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.0AASATTSRR44 pKa = 11.84TTRR47 pKa = 11.84KK48 pKa = 8.21ATPTSSKK55 pKa = 9.87PARR58 pKa = 11.84KK59 pKa = 8.82QVSKK63 pKa = 10.81RR64 pKa = 11.84RR65 pKa = 11.84TAAGRR70 pKa = 11.84SRR72 pKa = 11.84QRR74 pKa = 11.84RR75 pKa = 11.84AA76 pKa = 2.78
Molecular weight: 8.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2195810 |
21 |
4433 |
303.8 |
33.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.74 ± 0.034 | 0.789 ± 0.01 |
5.679 ± 0.026 | 5.706 ± 0.029 |
3.996 ± 0.019 | 8.234 ± 0.031 |
2.027 ± 0.015 | 5.895 ± 0.02 |
3.707 ± 0.02 | 10.013 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.564 ± 0.014 | 2.911 ± 0.018 |
4.883 ± 0.022 | 3.142 ± 0.017 |
6.512 ± 0.031 | 5.907 ± 0.025 |
5.337 ± 0.023 | 7.269 ± 0.023 |
1.313 ± 0.011 | 2.375 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
