
Xanthomarina gelatinilytica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Xanthomarina
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
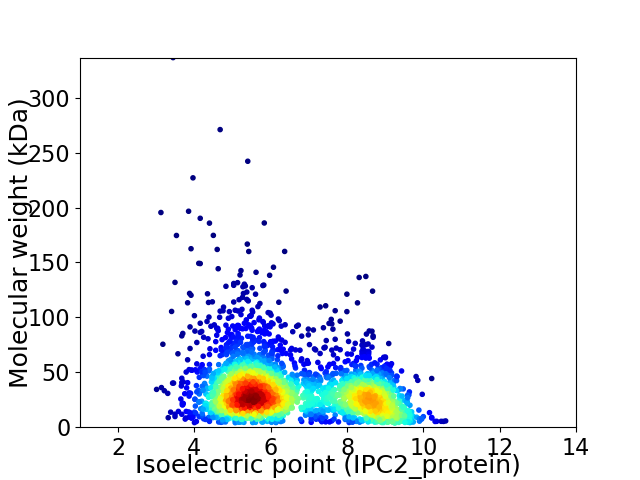
Virtual 2D-PAGE plot for 2841 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
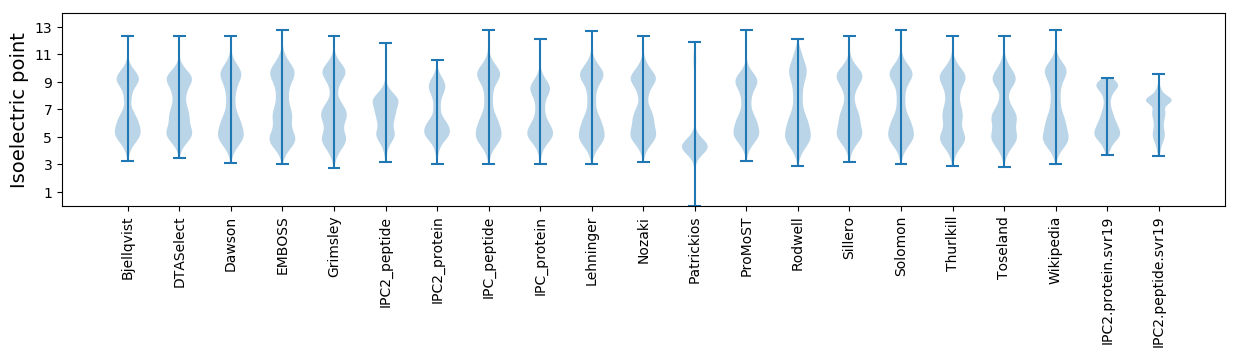
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M7N6T0|M7N6T0_9FLAO HTH cro/C1-type domain-containing protein OS=Xanthomarina gelatinilytica OX=1137281 GN=D778_01145 PE=3 SV=1
MM1 pKa = 7.47KK2 pKa = 10.4KK3 pKa = 9.73ILLTFLSFNAIIFAGFSQEE22 pKa = 3.67EE23 pKa = 4.67GFAIHH28 pKa = 6.31TVGPTGSNPYY38 pKa = 10.32VIDD41 pKa = 4.14SGHH44 pKa = 7.07LNPGAYY50 pKa = 8.79PDD52 pKa = 4.4LAVGTQSGNTVEE64 pKa = 5.12IYY66 pKa = 10.55INNEE70 pKa = 3.93DD71 pKa = 3.65GTFAAPIIKK80 pKa = 10.01NLSLVSGVHH89 pKa = 5.98IADD92 pKa = 3.08IDD94 pKa = 4.45GINGNDD100 pKa = 3.37VLASSSGGSKK110 pKa = 10.23LVWYY114 pKa = 9.97ANNGDD119 pKa = 3.69GTFADD124 pKa = 4.07EE125 pKa = 5.29AIISSSLDD133 pKa = 3.38TPGTIVTGYY142 pKa = 9.61IDD144 pKa = 4.34SDD146 pKa = 3.56ATLDD150 pKa = 3.51IVLSVYY156 pKa = 10.08GASGDD161 pKa = 3.54TDD163 pKa = 3.54RR164 pKa = 11.84VIWFANDD171 pKa = 3.36GLPWTEE177 pKa = 3.5QDD179 pKa = 4.05VVPATAGLGCGNIDD193 pKa = 3.58IADD196 pKa = 3.63VDD198 pKa = 4.18GDD200 pKa = 3.83TDD202 pKa = 4.83LDD204 pKa = 3.51IVVANTDD211 pKa = 3.01AGTVEE216 pKa = 5.4LYY218 pKa = 10.84YY219 pKa = 11.23NNYY222 pKa = 9.77NPLTDD227 pKa = 3.92NNPVSFTEE235 pKa = 4.21SAGGDD240 pKa = 2.85ISTGNTYY247 pKa = 10.95LFGLDD252 pKa = 3.89FGDD255 pKa = 4.13VNDD258 pKa = 4.94DD259 pKa = 3.42SFMDD263 pKa = 3.51IVKK266 pKa = 10.09VDD268 pKa = 3.85LSGHH272 pKa = 6.33EE273 pKa = 3.53IAYY276 pKa = 7.89YY277 pKa = 10.55QNDD280 pKa = 3.92GAGNFTEE287 pKa = 4.8VPVSSHH293 pKa = 5.8QYY295 pKa = 8.99PSIAFVTDD303 pKa = 4.1LNNDD307 pKa = 3.81GYY309 pKa = 11.48NDD311 pKa = 3.18IVAADD316 pKa = 4.25GLNQNDD322 pKa = 3.29DD323 pKa = 4.05MFWFEE328 pKa = 4.13SDD330 pKa = 3.08AFGGLGAEE338 pKa = 4.44TIITNNTLHH347 pKa = 5.78NQVYY351 pKa = 10.17NFTINDD357 pKa = 3.54FDD359 pKa = 4.42NDD361 pKa = 3.51GDD363 pKa = 3.93IDD365 pKa = 4.19FATLGYY371 pKa = 10.26QDD373 pKa = 4.99GTLKK377 pKa = 10.38WIEE380 pKa = 4.13NNLNLIGTQEE390 pKa = 4.04FKK392 pKa = 11.38LNTVSIYY399 pKa = 10.38PNPTSNRR406 pKa = 11.84IYY408 pKa = 10.83FKK410 pKa = 10.9GLTSNTEE417 pKa = 3.75RR418 pKa = 11.84VSVYY422 pKa = 10.62DD423 pKa = 3.45ILGKK427 pKa = 10.57KK428 pKa = 9.39ILSKK432 pKa = 9.74TVNTNKK438 pKa = 10.45GLDD441 pKa = 3.5VSKK444 pKa = 10.1LQNGIYY450 pKa = 9.79ILKK453 pKa = 9.96LDD455 pKa = 4.43QLNTSYY461 pKa = 11.5KK462 pKa = 9.75FVKK465 pKa = 10.24KK466 pKa = 10.63
MM1 pKa = 7.47KK2 pKa = 10.4KK3 pKa = 9.73ILLTFLSFNAIIFAGFSQEE22 pKa = 3.67EE23 pKa = 4.67GFAIHH28 pKa = 6.31TVGPTGSNPYY38 pKa = 10.32VIDD41 pKa = 4.14SGHH44 pKa = 7.07LNPGAYY50 pKa = 8.79PDD52 pKa = 4.4LAVGTQSGNTVEE64 pKa = 5.12IYY66 pKa = 10.55INNEE70 pKa = 3.93DD71 pKa = 3.65GTFAAPIIKK80 pKa = 10.01NLSLVSGVHH89 pKa = 5.98IADD92 pKa = 3.08IDD94 pKa = 4.45GINGNDD100 pKa = 3.37VLASSSGGSKK110 pKa = 10.23LVWYY114 pKa = 9.97ANNGDD119 pKa = 3.69GTFADD124 pKa = 4.07EE125 pKa = 5.29AIISSSLDD133 pKa = 3.38TPGTIVTGYY142 pKa = 9.61IDD144 pKa = 4.34SDD146 pKa = 3.56ATLDD150 pKa = 3.51IVLSVYY156 pKa = 10.08GASGDD161 pKa = 3.54TDD163 pKa = 3.54RR164 pKa = 11.84VIWFANDD171 pKa = 3.36GLPWTEE177 pKa = 3.5QDD179 pKa = 4.05VVPATAGLGCGNIDD193 pKa = 3.58IADD196 pKa = 3.63VDD198 pKa = 4.18GDD200 pKa = 3.83TDD202 pKa = 4.83LDD204 pKa = 3.51IVVANTDD211 pKa = 3.01AGTVEE216 pKa = 5.4LYY218 pKa = 10.84YY219 pKa = 11.23NNYY222 pKa = 9.77NPLTDD227 pKa = 3.92NNPVSFTEE235 pKa = 4.21SAGGDD240 pKa = 2.85ISTGNTYY247 pKa = 10.95LFGLDD252 pKa = 3.89FGDD255 pKa = 4.13VNDD258 pKa = 4.94DD259 pKa = 3.42SFMDD263 pKa = 3.51IVKK266 pKa = 10.09VDD268 pKa = 3.85LSGHH272 pKa = 6.33EE273 pKa = 3.53IAYY276 pKa = 7.89YY277 pKa = 10.55QNDD280 pKa = 3.92GAGNFTEE287 pKa = 4.8VPVSSHH293 pKa = 5.8QYY295 pKa = 8.99PSIAFVTDD303 pKa = 4.1LNNDD307 pKa = 3.81GYY309 pKa = 11.48NDD311 pKa = 3.18IVAADD316 pKa = 4.25GLNQNDD322 pKa = 3.29DD323 pKa = 4.05MFWFEE328 pKa = 4.13SDD330 pKa = 3.08AFGGLGAEE338 pKa = 4.44TIITNNTLHH347 pKa = 5.78NQVYY351 pKa = 10.17NFTINDD357 pKa = 3.54FDD359 pKa = 4.42NDD361 pKa = 3.51GDD363 pKa = 3.93IDD365 pKa = 4.19FATLGYY371 pKa = 10.26QDD373 pKa = 4.99GTLKK377 pKa = 10.38WIEE380 pKa = 4.13NNLNLIGTQEE390 pKa = 4.04FKK392 pKa = 11.38LNTVSIYY399 pKa = 10.38PNPTSNRR406 pKa = 11.84IYY408 pKa = 10.83FKK410 pKa = 10.9GLTSNTEE417 pKa = 3.75RR418 pKa = 11.84VSVYY422 pKa = 10.62DD423 pKa = 3.45ILGKK427 pKa = 10.57KK428 pKa = 9.39ILSKK432 pKa = 9.74TVNTNKK438 pKa = 10.45GLDD441 pKa = 3.5VSKK444 pKa = 10.1LQNGIYY450 pKa = 9.79ILKK453 pKa = 9.96LDD455 pKa = 4.43QLNTSYY461 pKa = 11.5KK462 pKa = 9.75FVKK465 pKa = 10.24KK466 pKa = 10.63
Molecular weight: 50.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7N096|M7N096_9FLAO Two-component system sensor histidine kinase/response OS=Xanthomarina gelatinilytica OX=1137281 GN=D778_02690 PE=4 SV=1
MM1 pKa = 8.08DD2 pKa = 5.78LLRR5 pKa = 11.84HH6 pKa = 5.76VIIVTKK12 pKa = 10.47NPNGSVCKK20 pKa = 10.35NYY22 pKa = 10.48NPNIRR27 pKa = 11.84NHH29 pKa = 5.38VKK31 pKa = 10.29RR32 pKa = 11.84NQKK35 pKa = 9.71ILIRR39 pKa = 11.84EE40 pKa = 4.11
MM1 pKa = 8.08DD2 pKa = 5.78LLRR5 pKa = 11.84HH6 pKa = 5.76VIIVTKK12 pKa = 10.47NPNGSVCKK20 pKa = 10.35NYY22 pKa = 10.48NPNIRR27 pKa = 11.84NHH29 pKa = 5.38VKK31 pKa = 10.29RR32 pKa = 11.84NQKK35 pKa = 9.71ILIRR39 pKa = 11.84EE40 pKa = 4.11
Molecular weight: 4.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
902307 |
37 |
3221 |
317.6 |
35.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.337 ± 0.044 | 0.761 ± 0.018 |
5.497 ± 0.034 | 6.52 ± 0.051 |
5.266 ± 0.034 | 6.13 ± 0.054 |
1.967 ± 0.022 | 7.877 ± 0.038 |
7.815 ± 0.068 | 9.442 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.262 ± 0.029 | 6.346 ± 0.057 |
3.377 ± 0.026 | 3.578 ± 0.032 |
3.087 ± 0.033 | 6.331 ± 0.037 |
5.986 ± 0.052 | 6.28 ± 0.038 |
0.989 ± 0.018 | 4.151 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
