
Granulicella pectinivorans
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Granulicella
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
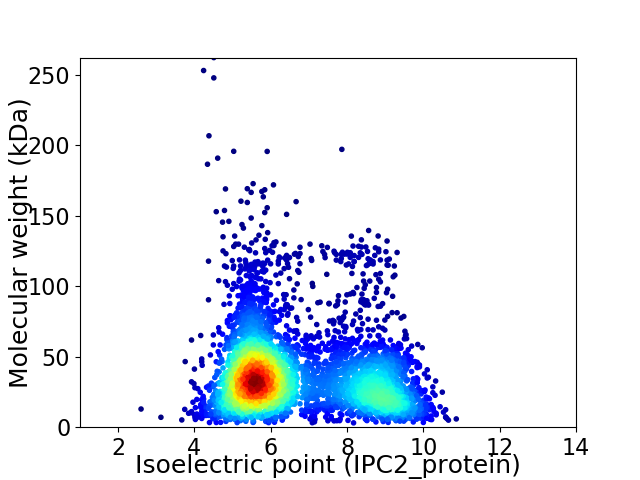
Virtual 2D-PAGE plot for 4259 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6L8V8|A0A1I6L8V8_9BACT Quinolinate phosphoribosyltransferase [decarboxylating] OS=Granulicella pectinivorans OX=474950 GN=SAMN05421771_0428 PE=3 SV=1
MM1 pKa = 7.51PEE3 pKa = 4.05EE4 pKa = 4.21VRR6 pKa = 11.84STTANQLVILFEE18 pKa = 4.37GSWFFIQDD26 pKa = 3.68PEE28 pKa = 4.46DD29 pKa = 3.39STRR32 pKa = 11.84ILALCPYY39 pKa = 9.83VDD41 pKa = 5.48APDD44 pKa = 3.72HH45 pKa = 5.78VCWFGFWDD53 pKa = 4.68GSSIAGPNATGPNGAMQEE71 pKa = 4.4GEE73 pKa = 4.61SLWVDD78 pKa = 4.16VITDD82 pKa = 3.58QAKK85 pKa = 8.38NTAPVDD91 pKa = 3.75ALFQAAQAAYY101 pKa = 9.58SFPYY105 pKa = 8.98LTSSDD110 pKa = 4.01PATSPLQLTDD120 pKa = 3.28TTGMRR125 pKa = 11.84RR126 pKa = 11.84VSISMPDD133 pKa = 3.16SMRR136 pKa = 11.84ADD138 pKa = 3.34GLLTNATVYY147 pKa = 10.75GSQNPGSTIKK157 pKa = 10.72LDD159 pKa = 3.73AGSSYY164 pKa = 11.5VDD166 pKa = 3.73FLFVYY171 pKa = 10.66YY172 pKa = 10.55YY173 pKa = 8.81DD174 pKa = 3.88TQADD178 pKa = 3.72VAMGTQFFEE187 pKa = 4.35QPVFSTSLATVVPHH201 pKa = 7.24LIFKK205 pKa = 10.75VMGTAADD212 pKa = 3.91AQDD215 pKa = 3.3MASEE219 pKa = 4.42YY220 pKa = 9.35THH222 pKa = 7.11LVQTFDD228 pKa = 3.88SLRR231 pKa = 11.84QMVFSPDD238 pKa = 4.39GICCDD243 pKa = 3.31VAIYY247 pKa = 10.28PEE249 pKa = 4.62AGQRR253 pKa = 11.84LQFEE257 pKa = 4.37ILDD260 pKa = 3.96PSFSAAEE267 pKa = 4.05LGIPSQQLGGLRR279 pKa = 11.84IRR281 pKa = 11.84DD282 pKa = 3.91YY283 pKa = 11.15ASCAAGPFAASGVLGGG299 pKa = 4.3
MM1 pKa = 7.51PEE3 pKa = 4.05EE4 pKa = 4.21VRR6 pKa = 11.84STTANQLVILFEE18 pKa = 4.37GSWFFIQDD26 pKa = 3.68PEE28 pKa = 4.46DD29 pKa = 3.39STRR32 pKa = 11.84ILALCPYY39 pKa = 9.83VDD41 pKa = 5.48APDD44 pKa = 3.72HH45 pKa = 5.78VCWFGFWDD53 pKa = 4.68GSSIAGPNATGPNGAMQEE71 pKa = 4.4GEE73 pKa = 4.61SLWVDD78 pKa = 4.16VITDD82 pKa = 3.58QAKK85 pKa = 8.38NTAPVDD91 pKa = 3.75ALFQAAQAAYY101 pKa = 9.58SFPYY105 pKa = 8.98LTSSDD110 pKa = 4.01PATSPLQLTDD120 pKa = 3.28TTGMRR125 pKa = 11.84RR126 pKa = 11.84VSISMPDD133 pKa = 3.16SMRR136 pKa = 11.84ADD138 pKa = 3.34GLLTNATVYY147 pKa = 10.75GSQNPGSTIKK157 pKa = 10.72LDD159 pKa = 3.73AGSSYY164 pKa = 11.5VDD166 pKa = 3.73FLFVYY171 pKa = 10.66YY172 pKa = 10.55YY173 pKa = 8.81DD174 pKa = 3.88TQADD178 pKa = 3.72VAMGTQFFEE187 pKa = 4.35QPVFSTSLATVVPHH201 pKa = 7.24LIFKK205 pKa = 10.75VMGTAADD212 pKa = 3.91AQDD215 pKa = 3.3MASEE219 pKa = 4.42YY220 pKa = 9.35THH222 pKa = 7.11LVQTFDD228 pKa = 3.88SLRR231 pKa = 11.84QMVFSPDD238 pKa = 4.39GICCDD243 pKa = 3.31VAIYY247 pKa = 10.28PEE249 pKa = 4.62AGQRR253 pKa = 11.84LQFEE257 pKa = 4.37ILDD260 pKa = 3.96PSFSAAEE267 pKa = 4.05LGIPSQQLGGLRR279 pKa = 11.84IRR281 pKa = 11.84DD282 pKa = 3.91YY283 pKa = 11.15ASCAAGPFAASGVLGGG299 pKa = 4.3
Molecular weight: 32.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6LHL1|A0A1I6LHL1_9BACT Uncharacterized protein OS=Granulicella pectinivorans OX=474950 GN=SAMN05421771_0711 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 5.69RR13 pKa = 11.84AKK15 pKa = 9.55THH17 pKa = 5.47GFLTRR22 pKa = 11.84MKK24 pKa = 9.03TKK26 pKa = 10.57AGAAVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.72KK42 pKa = 10.29IAVSAGFRR50 pKa = 11.84DD51 pKa = 3.66
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 5.69RR13 pKa = 11.84AKK15 pKa = 9.55THH17 pKa = 5.47GFLTRR22 pKa = 11.84MKK24 pKa = 9.03TKK26 pKa = 10.57AGAAVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.72KK42 pKa = 10.29IAVSAGFRR50 pKa = 11.84DD51 pKa = 3.66
Molecular weight: 5.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1581086 |
29 |
2642 |
371.2 |
40.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.182 ± 0.041 | 0.831 ± 0.011 |
4.992 ± 0.029 | 5.155 ± 0.047 |
3.918 ± 0.026 | 8.248 ± 0.045 |
2.267 ± 0.019 | 4.966 ± 0.03 |
3.691 ± 0.032 | 9.989 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.38 ± 0.019 | 3.274 ± 0.036 |
5.456 ± 0.026 | 3.645 ± 0.029 |
5.867 ± 0.043 | 6.121 ± 0.039 |
6.562 ± 0.056 | 7.363 ± 0.031 |
1.325 ± 0.016 | 2.768 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
