
Wad Medani virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
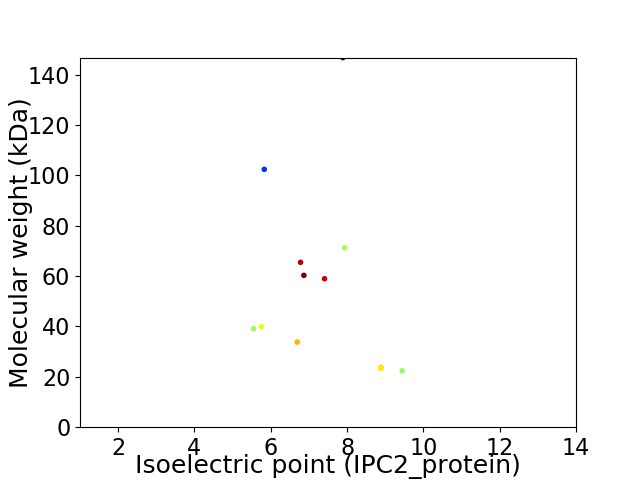
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4M910|A0A0H4M910_9REOV RNA-directed RNA polymerase OS=Wad Medani virus OX=40067 GN=VP1 PE=3 SV=1
MM1 pKa = 7.85DD2 pKa = 5.85AYY4 pKa = 10.27HH5 pKa = 6.97ARR7 pKa = 11.84ALTVLEE13 pKa = 4.25SLSMASEE20 pKa = 4.32ARR22 pKa = 11.84SHH24 pKa = 6.97RR25 pKa = 11.84DD26 pKa = 2.9PVTEE30 pKa = 3.95TTISIFAMRR39 pKa = 11.84FNATSNRR46 pKa = 11.84PITASPFTRR55 pKa = 11.84EE56 pKa = 3.37ARR58 pKa = 11.84RR59 pKa = 11.84NNFYY63 pKa = 10.97AALDD67 pKa = 3.59VTYY70 pKa = 10.51AALNITTDD78 pKa = 4.14FVLPDD83 pKa = 3.44YY84 pKa = 10.74SQNAQTLAILARR96 pKa = 11.84EE97 pKa = 4.1EE98 pKa = 4.36LPYY101 pKa = 10.74TPGSFRR107 pKa = 11.84RR108 pKa = 11.84MLRR111 pKa = 11.84IRR113 pKa = 11.84EE114 pKa = 4.27CTEE117 pKa = 3.23GAANVRR123 pKa = 11.84EE124 pKa = 4.21EE125 pKa = 4.21VDD127 pKa = 4.24PYY129 pKa = 9.53TQYY132 pKa = 11.25QIVVQTGCDD141 pKa = 4.0LDD143 pKa = 3.89TGAAGCAVFANSPNQLQVTIEE164 pKa = 3.87AGRR167 pKa = 11.84EE168 pKa = 3.57VDD170 pKa = 2.98ITADD174 pKa = 4.17LFPQDD179 pKa = 3.88RR180 pKa = 11.84EE181 pKa = 4.32VIAIEE186 pKa = 3.98VTVQILSHH194 pKa = 6.68AFHH197 pKa = 6.98NGAVMAHH204 pKa = 5.26EE205 pKa = 5.07RR206 pKa = 11.84ALEE209 pKa = 3.89MAIDD213 pKa = 4.31GMPLALGTRR222 pKa = 11.84VTVSVGSRR230 pKa = 11.84VTVANRR236 pKa = 11.84GVQNRR241 pKa = 11.84GIFVVTCHH249 pKa = 6.48RR250 pKa = 11.84FWVPDD255 pKa = 4.17PPRR258 pKa = 11.84IIYY261 pKa = 8.12DD262 pKa = 3.49TMEE265 pKa = 5.34ADD267 pKa = 2.97ILAVYY272 pKa = 8.44TYY274 pKa = 10.98KK275 pKa = 11.07DD276 pKa = 3.88RR277 pKa = 11.84VWDD280 pKa = 3.83ALRR283 pKa = 11.84AYY285 pKa = 10.11VLAAVGLPEE294 pKa = 4.06RR295 pKa = 11.84HH296 pKa = 6.08FPLTPVANPRR306 pKa = 11.84QVLAIALLSRR316 pKa = 11.84LFDD319 pKa = 3.86VYY321 pKa = 10.91CATHH325 pKa = 7.17PEE327 pKa = 4.21LALPMAVPGGALAQRR342 pKa = 11.84LAAALQVFRR351 pKa = 11.84DD352 pKa = 3.73ARR354 pKa = 3.73
MM1 pKa = 7.85DD2 pKa = 5.85AYY4 pKa = 10.27HH5 pKa = 6.97ARR7 pKa = 11.84ALTVLEE13 pKa = 4.25SLSMASEE20 pKa = 4.32ARR22 pKa = 11.84SHH24 pKa = 6.97RR25 pKa = 11.84DD26 pKa = 2.9PVTEE30 pKa = 3.95TTISIFAMRR39 pKa = 11.84FNATSNRR46 pKa = 11.84PITASPFTRR55 pKa = 11.84EE56 pKa = 3.37ARR58 pKa = 11.84RR59 pKa = 11.84NNFYY63 pKa = 10.97AALDD67 pKa = 3.59VTYY70 pKa = 10.51AALNITTDD78 pKa = 4.14FVLPDD83 pKa = 3.44YY84 pKa = 10.74SQNAQTLAILARR96 pKa = 11.84EE97 pKa = 4.1EE98 pKa = 4.36LPYY101 pKa = 10.74TPGSFRR107 pKa = 11.84RR108 pKa = 11.84MLRR111 pKa = 11.84IRR113 pKa = 11.84EE114 pKa = 4.27CTEE117 pKa = 3.23GAANVRR123 pKa = 11.84EE124 pKa = 4.21EE125 pKa = 4.21VDD127 pKa = 4.24PYY129 pKa = 9.53TQYY132 pKa = 11.25QIVVQTGCDD141 pKa = 4.0LDD143 pKa = 3.89TGAAGCAVFANSPNQLQVTIEE164 pKa = 3.87AGRR167 pKa = 11.84EE168 pKa = 3.57VDD170 pKa = 2.98ITADD174 pKa = 4.17LFPQDD179 pKa = 3.88RR180 pKa = 11.84EE181 pKa = 4.32VIAIEE186 pKa = 3.98VTVQILSHH194 pKa = 6.68AFHH197 pKa = 6.98NGAVMAHH204 pKa = 5.26EE205 pKa = 5.07RR206 pKa = 11.84ALEE209 pKa = 3.89MAIDD213 pKa = 4.31GMPLALGTRR222 pKa = 11.84VTVSVGSRR230 pKa = 11.84VTVANRR236 pKa = 11.84GVQNRR241 pKa = 11.84GIFVVTCHH249 pKa = 6.48RR250 pKa = 11.84FWVPDD255 pKa = 4.17PPRR258 pKa = 11.84IIYY261 pKa = 8.12DD262 pKa = 3.49TMEE265 pKa = 5.34ADD267 pKa = 2.97ILAVYY272 pKa = 8.44TYY274 pKa = 10.98KK275 pKa = 11.07DD276 pKa = 3.88RR277 pKa = 11.84VWDD280 pKa = 3.83ALRR283 pKa = 11.84AYY285 pKa = 10.11VLAAVGLPEE294 pKa = 4.06RR295 pKa = 11.84HH296 pKa = 6.08FPLTPVANPRR306 pKa = 11.84QVLAIALLSRR316 pKa = 11.84LFDD319 pKa = 3.86VYY321 pKa = 10.91CATHH325 pKa = 7.17PEE327 pKa = 4.21LALPMAVPGGALAQRR342 pKa = 11.84LAAALQVFRR351 pKa = 11.84DD352 pKa = 3.73ARR354 pKa = 3.73
Molecular weight: 39.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4MK95|A0A0H4MK95_9REOV NS4 protein OS=Wad Medani virus OX=40067 GN=NS4 PE=4 SV=1
MM1 pKa = 7.19MGSTMSQVTRR11 pKa = 11.84GALRR15 pKa = 11.84KK16 pKa = 9.37IEE18 pKa = 3.96RR19 pKa = 11.84RR20 pKa = 11.84LDD22 pKa = 3.98GIEE25 pKa = 3.93NFFEE29 pKa = 4.77NIHH32 pKa = 6.44PEE34 pKa = 4.35GEE36 pKa = 4.25GMAGEE41 pKa = 4.07IAEE44 pKa = 4.4NVPVEE49 pKa = 4.57DD50 pKa = 4.15PLALEE55 pKa = 4.5VLTSLEE61 pKa = 4.04WAVDD65 pKa = 3.54QMKK68 pKa = 10.18EE69 pKa = 3.65MCPRR73 pKa = 11.84IILNTGIRR81 pKa = 11.84VLEE84 pKa = 4.24KK85 pKa = 10.58RR86 pKa = 11.84PEE88 pKa = 4.17TPVRR92 pKa = 11.84MIHH95 pKa = 5.63VLAEE99 pKa = 3.95IRR101 pKa = 11.84WRR103 pKa = 11.84RR104 pKa = 11.84VMWTMLSIWLRR115 pKa = 11.84RR116 pKa = 11.84WIRR119 pKa = 11.84TPAGAYY125 pKa = 9.99LPSQNQLWVRR135 pKa = 11.84CARR138 pKa = 11.84ADD140 pKa = 3.33RR141 pKa = 11.84FMFSCLGIRR150 pKa = 11.84NHH152 pKa = 5.67VMEE155 pKa = 5.43MYY157 pKa = 10.68AIMSVRR163 pKa = 11.84RR164 pKa = 11.84CLKK167 pKa = 10.58LSDD170 pKa = 3.27CRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84LRR178 pKa = 11.84RR179 pKa = 11.84KK180 pKa = 7.27MRR182 pKa = 11.84RR183 pKa = 11.84LIGEE187 pKa = 4.31SNN189 pKa = 3.09
MM1 pKa = 7.19MGSTMSQVTRR11 pKa = 11.84GALRR15 pKa = 11.84KK16 pKa = 9.37IEE18 pKa = 3.96RR19 pKa = 11.84RR20 pKa = 11.84LDD22 pKa = 3.98GIEE25 pKa = 3.93NFFEE29 pKa = 4.77NIHH32 pKa = 6.44PEE34 pKa = 4.35GEE36 pKa = 4.25GMAGEE41 pKa = 4.07IAEE44 pKa = 4.4NVPVEE49 pKa = 4.57DD50 pKa = 4.15PLALEE55 pKa = 4.5VLTSLEE61 pKa = 4.04WAVDD65 pKa = 3.54QMKK68 pKa = 10.18EE69 pKa = 3.65MCPRR73 pKa = 11.84IILNTGIRR81 pKa = 11.84VLEE84 pKa = 4.24KK85 pKa = 10.58RR86 pKa = 11.84PEE88 pKa = 4.17TPVRR92 pKa = 11.84MIHH95 pKa = 5.63VLAEE99 pKa = 3.95IRR101 pKa = 11.84WRR103 pKa = 11.84RR104 pKa = 11.84VMWTMLSIWLRR115 pKa = 11.84RR116 pKa = 11.84WIRR119 pKa = 11.84TPAGAYY125 pKa = 9.99LPSQNQLWVRR135 pKa = 11.84CARR138 pKa = 11.84ADD140 pKa = 3.33RR141 pKa = 11.84FMFSCLGIRR150 pKa = 11.84NHH152 pKa = 5.67VMEE155 pKa = 5.43MYY157 pKa = 10.68AIMSVRR163 pKa = 11.84RR164 pKa = 11.84CLKK167 pKa = 10.58LSDD170 pKa = 3.27CRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84LRR178 pKa = 11.84RR179 pKa = 11.84KK180 pKa = 7.27MRR182 pKa = 11.84RR183 pKa = 11.84LIGEE187 pKa = 4.31SNN189 pKa = 3.09
Molecular weight: 22.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6156 |
189 |
1303 |
513.0 |
57.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.006 ± 0.672 | 1.413 ± 0.323 |
5.799 ± 0.372 | 5.491 ± 0.291 |
3.46 ± 0.339 | 5.962 ± 0.315 |
2.648 ± 0.201 | 4.906 ± 0.337 |
3.054 ± 0.404 | 9.649 ± 0.438 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.226 | 2.875 ± 0.141 |
5.75 ± 0.396 | 3.265 ± 0.246 |
8.707 ± 0.406 | 7.001 ± 0.569 |
5.962 ± 0.382 | 6.774 ± 0.361 |
1.381 ± 0.235 | 3.005 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
