
Clostridium sp. CAG:127
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
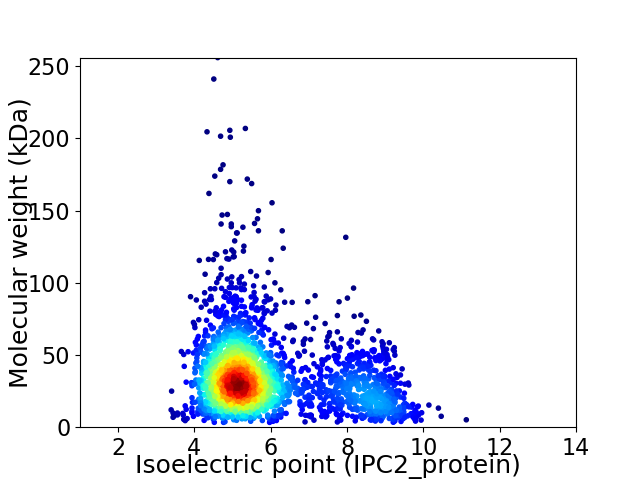
Virtual 2D-PAGE plot for 2463 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
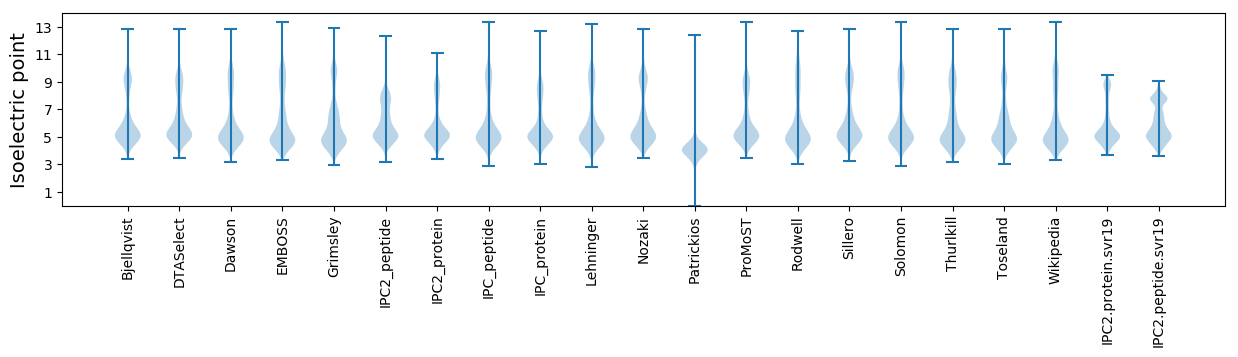
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5PC08|R5PC08_9CLOT Capsular biosynthesis protein OS=Clostridium sp. CAG:127 OX=1262774 GN=BN482_00830 PE=4 SV=1
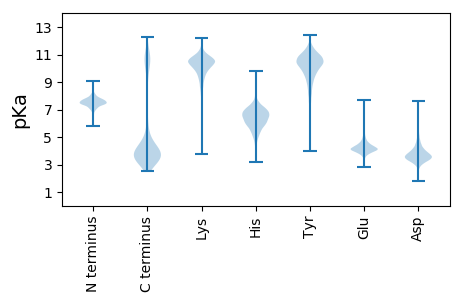
MM1 pKa = 7.36KK2 pKa = 10.22VCEE5 pKa = 4.08MCGAQNDD12 pKa = 4.23DD13 pKa = 3.81VFTKK17 pKa = 10.66CSVCGADD24 pKa = 3.79LPSMNSQDD32 pKa = 3.74GDD34 pKa = 3.78DD35 pKa = 4.32EE36 pKa = 4.49KK37 pKa = 10.75TVLIDD42 pKa = 3.8PDD44 pKa = 3.5QAPAQPSGLSLKK56 pKa = 9.72TDD58 pKa = 3.31TSTYY62 pKa = 9.05GQADD66 pKa = 3.69AGQTNPQPAAPQFGGMGQPQMNGMGQPQMNGMGQPGMNGMPQQGMPGQAPYY117 pKa = 9.93GQPGFGGPVPAQPAKK132 pKa = 9.81PKK134 pKa = 9.91KK135 pKa = 10.06GLIIGIIAAAVVVLAVAFVLIFKK158 pKa = 10.52GGFGAKK164 pKa = 9.72GGASSPEE171 pKa = 3.51AVAQQFIEE179 pKa = 4.11AMNDD183 pKa = 2.93QDD185 pKa = 4.0VDD187 pKa = 3.96KK188 pKa = 10.38MAALCPPFLDD198 pKa = 4.74PGQEE202 pKa = 4.45DD203 pKa = 3.92IEE205 pKa = 5.05DD206 pKa = 3.85MLDD209 pKa = 3.32SYY211 pKa = 11.55SGYY214 pKa = 10.84DD215 pKa = 3.06VTFKK219 pKa = 11.54YY220 pKa = 10.46EE221 pKa = 4.14GVEE224 pKa = 4.12SKK226 pKa = 10.9EE227 pKa = 4.37MYY229 pKa = 10.69DD230 pKa = 5.54SDD232 pKa = 5.64DD233 pKa = 4.83LDD235 pKa = 4.88DD236 pKa = 5.15LEE238 pKa = 5.53EE239 pKa = 5.72DD240 pKa = 3.24ISDD243 pKa = 4.07YY244 pKa = 11.46ADD246 pKa = 3.37KK247 pKa = 10.63TIKK250 pKa = 10.76LQDD253 pKa = 3.56ACDD256 pKa = 3.86LEE258 pKa = 5.24FSFNVAVTMSGEE270 pKa = 4.42TYY272 pKa = 9.96DD273 pKa = 3.86QSSTYY278 pKa = 10.38PITCIKK284 pKa = 10.81YY285 pKa = 8.6KK286 pKa = 10.72GNWYY290 pKa = 10.2LYY292 pKa = 9.61EE293 pKa = 4.06
MM1 pKa = 7.36KK2 pKa = 10.22VCEE5 pKa = 4.08MCGAQNDD12 pKa = 4.23DD13 pKa = 3.81VFTKK17 pKa = 10.66CSVCGADD24 pKa = 3.79LPSMNSQDD32 pKa = 3.74GDD34 pKa = 3.78DD35 pKa = 4.32EE36 pKa = 4.49KK37 pKa = 10.75TVLIDD42 pKa = 3.8PDD44 pKa = 3.5QAPAQPSGLSLKK56 pKa = 9.72TDD58 pKa = 3.31TSTYY62 pKa = 9.05GQADD66 pKa = 3.69AGQTNPQPAAPQFGGMGQPQMNGMGQPQMNGMGQPGMNGMPQQGMPGQAPYY117 pKa = 9.93GQPGFGGPVPAQPAKK132 pKa = 9.81PKK134 pKa = 9.91KK135 pKa = 10.06GLIIGIIAAAVVVLAVAFVLIFKK158 pKa = 10.52GGFGAKK164 pKa = 9.72GGASSPEE171 pKa = 3.51AVAQQFIEE179 pKa = 4.11AMNDD183 pKa = 2.93QDD185 pKa = 4.0VDD187 pKa = 3.96KK188 pKa = 10.38MAALCPPFLDD198 pKa = 4.74PGQEE202 pKa = 4.45DD203 pKa = 3.92IEE205 pKa = 5.05DD206 pKa = 3.85MLDD209 pKa = 3.32SYY211 pKa = 11.55SGYY214 pKa = 10.84DD215 pKa = 3.06VTFKK219 pKa = 11.54YY220 pKa = 10.46EE221 pKa = 4.14GVEE224 pKa = 4.12SKK226 pKa = 10.9EE227 pKa = 4.37MYY229 pKa = 10.69DD230 pKa = 5.54SDD232 pKa = 5.64DD233 pKa = 4.83LDD235 pKa = 4.88DD236 pKa = 5.15LEE238 pKa = 5.53EE239 pKa = 5.72DD240 pKa = 3.24ISDD243 pKa = 4.07YY244 pKa = 11.46ADD246 pKa = 3.37KK247 pKa = 10.63TIKK250 pKa = 10.76LQDD253 pKa = 3.56ACDD256 pKa = 3.86LEE258 pKa = 5.24FSFNVAVTMSGEE270 pKa = 4.42TYY272 pKa = 9.96DD273 pKa = 3.86QSSTYY278 pKa = 10.38PITCIKK284 pKa = 10.81YY285 pKa = 8.6KK286 pKa = 10.72GNWYY290 pKa = 10.2LYY292 pKa = 9.61EE293 pKa = 4.06
Molecular weight: 31.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5NU32|R5NU32_9CLOT Putative tRNA (cytidine(34)-2'-O)-methyltransferase OS=Clostridium sp. CAG:127 OX=1262774 GN=BN482_00769 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
812000 |
29 |
2354 |
329.7 |
36.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.779 ± 0.052 | 1.46 ± 0.021 |
6.129 ± 0.047 | 7.393 ± 0.058 |
3.845 ± 0.034 | 6.788 ± 0.051 |
1.74 ± 0.021 | 7.397 ± 0.041 |
6.892 ± 0.041 | 8.54 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.216 ± 0.026 | 4.496 ± 0.037 |
3.05 ± 0.026 | 3.433 ± 0.027 |
4.095 ± 0.036 | 5.527 ± 0.039 |
5.726 ± 0.053 | 7.191 ± 0.04 |
0.801 ± 0.016 | 4.502 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
