
Sphaerobacter thermophilus (strain DSM 20745 / S 6022)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Thermomicrobia; Sphaerobacteridae; Sphaerobacterales; Sphaerobacterineae; Sphaerobacteraceae; Sphaerobacter; Sphaerobacter thermophilus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
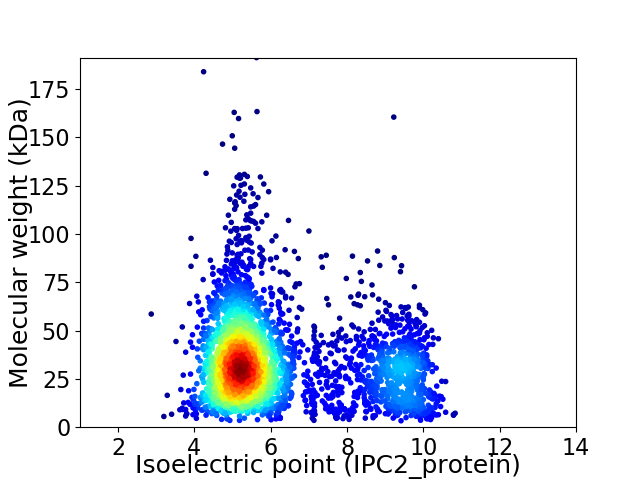
Virtual 2D-PAGE plot for 3471 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D1CAD0|D1CAD0_SPHTD Zeaxanthin epoxidase OS=Sphaerobacter thermophilus (strain DSM 20745 / S 6022) OX=479434 GN=Sthe_3373 PE=4 SV=1
MM1 pKa = 7.42RR2 pKa = 11.84TRR4 pKa = 11.84LVRR7 pKa = 11.84LLVILALMLSTMTSLGFAAAQQAGNPSTSMPGRR40 pKa = 11.84YY41 pKa = 8.6LVALKK46 pKa = 9.53STGDD50 pKa = 3.4ARR52 pKa = 11.84FMQAQITEE60 pKa = 4.05MGATVVQNLAPLNLLVVHH78 pKa = 6.25GTEE81 pKa = 4.55QVKK84 pKa = 9.87QRR86 pKa = 11.84LKK88 pKa = 10.65ADD90 pKa = 3.5PRR92 pKa = 11.84VSGVATDD99 pKa = 4.59RR100 pKa = 11.84IQRR103 pKa = 11.84IVPPEE108 pKa = 4.02LAPDD112 pKa = 3.87MFSSPQAAEE121 pKa = 4.82PIQIDD126 pKa = 4.12LTDD129 pKa = 4.17DD130 pKa = 3.76AQNGEE135 pKa = 4.27ITGDD139 pKa = 3.62PAFDD143 pKa = 4.64LSGLMWNIDD152 pKa = 3.86RR153 pKa = 11.84IEE155 pKa = 5.13APDD158 pKa = 3.2AWEE161 pKa = 4.24TTSGEE166 pKa = 4.15PEE168 pKa = 4.36VLVGVADD175 pKa = 3.82TGLDD179 pKa = 3.53STHH182 pKa = 6.84VDD184 pKa = 3.51LEE186 pKa = 4.7DD187 pKa = 4.69NIHH190 pKa = 5.96SVVDD194 pKa = 3.92FTEE197 pKa = 6.05AEE199 pKa = 4.43DD200 pKa = 4.9PPICDD205 pKa = 2.98TFFGVSDD212 pKa = 3.64QALAAQFGGPADD224 pKa = 4.28GDD226 pKa = 4.28WNGHH230 pKa = 5.07GSWIGGNIAAVLNEE244 pKa = 3.73QGVNGVAPNVKK255 pKa = 9.85LVSLKK260 pKa = 9.75ISQWCGYY267 pKa = 10.28AYY269 pKa = 10.62DD270 pKa = 4.03STILSAFLYY279 pKa = 10.36AASNDD284 pKa = 3.08IDD286 pKa = 4.07VVSLSFGGYY295 pKa = 9.6LDD297 pKa = 5.06RR298 pKa = 11.84SDD300 pKa = 4.61PEE302 pKa = 3.36QDD304 pKa = 4.45LIYY307 pKa = 10.37SQYY310 pKa = 11.28ASAVALARR318 pKa = 11.84SEE320 pKa = 4.08GTVIVAAAGNEE331 pKa = 3.98HH332 pKa = 7.17VEE334 pKa = 4.23IGDD337 pKa = 3.76GGQVLSSGMLTTPGTLLDD355 pKa = 4.7GFLDD359 pKa = 3.84LQGLYY364 pKa = 9.09EE365 pKa = 4.45IPGGIPGVVTVGSTGNMVNEE385 pKa = 4.73PSDD388 pKa = 3.76EE389 pKa = 4.27CPEE392 pKa = 4.15GTTEE396 pKa = 5.71GSLATCKK403 pKa = 10.67PEE405 pKa = 5.42DD406 pKa = 4.95DD407 pKa = 3.41PHH409 pKa = 7.51QPFGAGEE416 pKa = 4.06EE417 pKa = 4.2NQLAYY422 pKa = 10.26YY423 pKa = 10.29SNYY426 pKa = 9.53GPRR429 pKa = 11.84IDD431 pKa = 5.05LVAPGGSRR439 pKa = 11.84MFNLPLWDD447 pKa = 3.81RR448 pKa = 11.84GGTPGFPYY456 pKa = 10.16TIEE459 pKa = 4.32DD460 pKa = 5.15EE461 pKa = 4.37YY462 pKa = 11.19TAWQTFSTTSNWALYY477 pKa = 7.64TAQIPCFLFIGGGFPAGQCYY497 pKa = 7.21TTIQGTSMATPHH509 pKa = 5.71VSAVLALVASANPDD523 pKa = 3.25LRR525 pKa = 11.84GDD527 pKa = 3.59PDD529 pKa = 3.71RR530 pKa = 11.84LVEE533 pKa = 4.2IVKK536 pKa = 8.6EE537 pKa = 4.1TAQPISGNEE546 pKa = 4.01TPPLDD551 pKa = 4.87PSDD554 pKa = 4.13TSPGDD559 pKa = 3.06IGGPSCPTGYY569 pKa = 10.1CHH571 pKa = 7.77LGGDD575 pKa = 4.99PIPDD579 pKa = 3.32EE580 pKa = 4.43DD581 pKa = 4.62AYY583 pKa = 11.0GAGLVDD589 pKa = 3.81AAAAVEE595 pKa = 3.93AAMVEE600 pKa = 4.39DD601 pKa = 4.1EE602 pKa = 5.7AGSQSQPP609 pKa = 3.01
MM1 pKa = 7.42RR2 pKa = 11.84TRR4 pKa = 11.84LVRR7 pKa = 11.84LLVILALMLSTMTSLGFAAAQQAGNPSTSMPGRR40 pKa = 11.84YY41 pKa = 8.6LVALKK46 pKa = 9.53STGDD50 pKa = 3.4ARR52 pKa = 11.84FMQAQITEE60 pKa = 4.05MGATVVQNLAPLNLLVVHH78 pKa = 6.25GTEE81 pKa = 4.55QVKK84 pKa = 9.87QRR86 pKa = 11.84LKK88 pKa = 10.65ADD90 pKa = 3.5PRR92 pKa = 11.84VSGVATDD99 pKa = 4.59RR100 pKa = 11.84IQRR103 pKa = 11.84IVPPEE108 pKa = 4.02LAPDD112 pKa = 3.87MFSSPQAAEE121 pKa = 4.82PIQIDD126 pKa = 4.12LTDD129 pKa = 4.17DD130 pKa = 3.76AQNGEE135 pKa = 4.27ITGDD139 pKa = 3.62PAFDD143 pKa = 4.64LSGLMWNIDD152 pKa = 3.86RR153 pKa = 11.84IEE155 pKa = 5.13APDD158 pKa = 3.2AWEE161 pKa = 4.24TTSGEE166 pKa = 4.15PEE168 pKa = 4.36VLVGVADD175 pKa = 3.82TGLDD179 pKa = 3.53STHH182 pKa = 6.84VDD184 pKa = 3.51LEE186 pKa = 4.7DD187 pKa = 4.69NIHH190 pKa = 5.96SVVDD194 pKa = 3.92FTEE197 pKa = 6.05AEE199 pKa = 4.43DD200 pKa = 4.9PPICDD205 pKa = 2.98TFFGVSDD212 pKa = 3.64QALAAQFGGPADD224 pKa = 4.28GDD226 pKa = 4.28WNGHH230 pKa = 5.07GSWIGGNIAAVLNEE244 pKa = 3.73QGVNGVAPNVKK255 pKa = 9.85LVSLKK260 pKa = 9.75ISQWCGYY267 pKa = 10.28AYY269 pKa = 10.62DD270 pKa = 4.03STILSAFLYY279 pKa = 10.36AASNDD284 pKa = 3.08IDD286 pKa = 4.07VVSLSFGGYY295 pKa = 9.6LDD297 pKa = 5.06RR298 pKa = 11.84SDD300 pKa = 4.61PEE302 pKa = 3.36QDD304 pKa = 4.45LIYY307 pKa = 10.37SQYY310 pKa = 11.28ASAVALARR318 pKa = 11.84SEE320 pKa = 4.08GTVIVAAAGNEE331 pKa = 3.98HH332 pKa = 7.17VEE334 pKa = 4.23IGDD337 pKa = 3.76GGQVLSSGMLTTPGTLLDD355 pKa = 4.7GFLDD359 pKa = 3.84LQGLYY364 pKa = 9.09EE365 pKa = 4.45IPGGIPGVVTVGSTGNMVNEE385 pKa = 4.73PSDD388 pKa = 3.76EE389 pKa = 4.27CPEE392 pKa = 4.15GTTEE396 pKa = 5.71GSLATCKK403 pKa = 10.67PEE405 pKa = 5.42DD406 pKa = 4.95DD407 pKa = 3.41PHH409 pKa = 7.51QPFGAGEE416 pKa = 4.06EE417 pKa = 4.2NQLAYY422 pKa = 10.26YY423 pKa = 10.29SNYY426 pKa = 9.53GPRR429 pKa = 11.84IDD431 pKa = 5.05LVAPGGSRR439 pKa = 11.84MFNLPLWDD447 pKa = 3.81RR448 pKa = 11.84GGTPGFPYY456 pKa = 10.16TIEE459 pKa = 4.32DD460 pKa = 5.15EE461 pKa = 4.37YY462 pKa = 11.19TAWQTFSTTSNWALYY477 pKa = 7.64TAQIPCFLFIGGGFPAGQCYY497 pKa = 7.21TTIQGTSMATPHH509 pKa = 5.71VSAVLALVASANPDD523 pKa = 3.25LRR525 pKa = 11.84GDD527 pKa = 3.59PDD529 pKa = 3.71RR530 pKa = 11.84LVEE533 pKa = 4.2IVKK536 pKa = 8.6EE537 pKa = 4.1TAQPISGNEE546 pKa = 4.01TPPLDD551 pKa = 4.87PSDD554 pKa = 4.13TSPGDD559 pKa = 3.06IGGPSCPTGYY569 pKa = 10.1CHH571 pKa = 7.77LGGDD575 pKa = 4.99PIPDD579 pKa = 3.32EE580 pKa = 4.43DD581 pKa = 4.62AYY583 pKa = 11.0GAGLVDD589 pKa = 3.81AAAAVEE595 pKa = 3.93AAMVEE600 pKa = 4.39DD601 pKa = 4.1EE602 pKa = 5.7AGSQSQPP609 pKa = 3.01
Molecular weight: 63.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D1C246|D1C246_SPHTD Gas vesicle G OS=Sphaerobacter thermophilus (strain DSM 20745 / S 6022) OX=479434 GN=Sthe_0876 PE=4 SV=1
MM1 pKa = 7.28NRR3 pKa = 11.84RR4 pKa = 11.84SRR6 pKa = 11.84QILALTAAALATWTFPFNQVAAIGWLTVAILILPTPRR43 pKa = 11.84LKK45 pKa = 10.93PEE47 pKa = 3.72VVPRR51 pKa = 11.84QRR53 pKa = 11.84LRR55 pKa = 11.84NFLRR59 pKa = 11.84RR60 pKa = 11.84FRR62 pKa = 4.73
MM1 pKa = 7.28NRR3 pKa = 11.84RR4 pKa = 11.84SRR6 pKa = 11.84QILALTAAALATWTFPFNQVAAIGWLTVAILILPTPRR43 pKa = 11.84LKK45 pKa = 10.93PEE47 pKa = 3.72VVPRR51 pKa = 11.84QRR53 pKa = 11.84LRR55 pKa = 11.84NFLRR59 pKa = 11.84RR60 pKa = 11.84FRR62 pKa = 4.73
Molecular weight: 7.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1135642 |
31 |
1776 |
327.2 |
35.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.136 ± 0.057 | 0.679 ± 0.011 |
5.617 ± 0.032 | 6.397 ± 0.041 |
3.135 ± 0.021 | 8.57 ± 0.038 |
2.05 ± 0.018 | 4.973 ± 0.029 |
1.67 ± 0.022 | 10.546 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.99 ± 0.017 | 2.057 ± 0.02 |
6.036 ± 0.035 | 3.172 ± 0.025 |
8.355 ± 0.044 | 4.511 ± 0.025 |
5.619 ± 0.036 | 8.545 ± 0.035 |
1.466 ± 0.02 | 2.476 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
