
Oleiphilus messinensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oleiphilaceae; Oleiphilus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
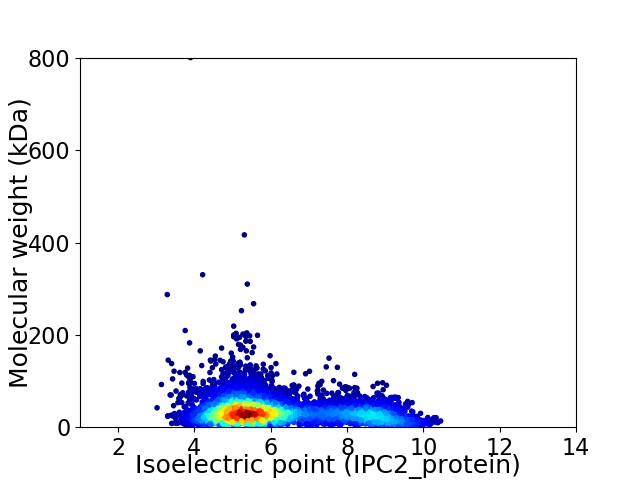
Virtual 2D-PAGE plot for 5456 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y0I9X1|A0A1Y0I9X1_9GAMM Uncharacterized protein OS=Oleiphilus messinensis OX=141451 GN=OLMES_2153 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84PSRR5 pKa = 11.84SLLVDD10 pKa = 5.0PISGPSHH17 pKa = 6.29GKK19 pKa = 10.09ALLSDD24 pKa = 3.68TLLRR28 pKa = 11.84AQNALQDD35 pKa = 4.21FVAKK39 pKa = 10.48DD40 pKa = 3.54VFDD43 pKa = 3.82TKK45 pKa = 11.07VFTVAFGEE53 pKa = 4.23NYY55 pKa = 10.17DD56 pKa = 4.14AIIFEE61 pKa = 4.45SLIDD65 pKa = 3.43QYY67 pKa = 11.95RR68 pKa = 11.84NGNFSSLPSIEE79 pKa = 4.27LRR81 pKa = 11.84SGLEE85 pKa = 3.73LQGADD90 pKa = 3.15GVYY93 pKa = 10.09VEE95 pKa = 4.85SLNAIFISDD104 pKa = 3.91TFYY107 pKa = 11.88GDD109 pKa = 4.88ANDD112 pKa = 4.07TQQVAVVLEE121 pKa = 4.64EE122 pKa = 3.9IGHH125 pKa = 7.02AIDD128 pKa = 4.74ALINKK133 pKa = 9.21SDD135 pKa = 3.61SKK137 pKa = 11.22GDD139 pKa = 3.42EE140 pKa = 3.59GRR142 pKa = 11.84IFAKK146 pKa = 10.45LVLGQEE152 pKa = 4.14MSDD155 pKa = 3.33EE156 pKa = 4.57VVAEE160 pKa = 3.99LHH162 pKa = 7.16AEE164 pKa = 4.17DD165 pKa = 5.9DD166 pKa = 4.31SLTLTVDD173 pKa = 3.51GQTVLAEE180 pKa = 4.35ASTKK184 pKa = 10.18VWDD187 pKa = 4.01SGDD190 pKa = 3.01IGGPYY195 pKa = 8.44ATTWKK200 pKa = 10.48YY201 pKa = 10.24ILKK204 pKa = 9.39TIDD207 pKa = 3.31TPYY210 pKa = 11.17SMSSTYY216 pKa = 10.37FEE218 pKa = 4.21NTLKK222 pKa = 10.44HH223 pKa = 6.19DD224 pKa = 4.06RR225 pKa = 11.84TWSDD229 pKa = 3.57FIGNKK234 pKa = 6.94TKK236 pKa = 10.29YY237 pKa = 8.24YY238 pKa = 10.23EE239 pKa = 4.18YY240 pKa = 10.35YY241 pKa = 10.97GEE243 pKa = 4.38VNGTSGDD250 pKa = 4.09DD251 pKa = 3.73IIIGATDD258 pKa = 3.53TKK260 pKa = 11.0KK261 pKa = 10.44KK262 pKa = 10.07WEE264 pKa = 4.91DD265 pKa = 3.58GNNDD269 pKa = 2.43IYY271 pKa = 11.13TWLRR275 pKa = 11.84AQEE278 pKa = 3.88EE279 pKa = 4.9TINGGSGDD287 pKa = 4.24DD288 pKa = 4.42YY289 pKa = 11.46IDD291 pKa = 4.41GGDD294 pKa = 4.08GSDD297 pKa = 4.32TINGGAGNDD306 pKa = 4.09LIFTGDD312 pKa = 3.85LSDD315 pKa = 5.17GSDD318 pKa = 3.7DD319 pKa = 4.69DD320 pKa = 5.4VITGGSGADD329 pKa = 3.2TFYY332 pKa = 10.74TGIITHH338 pKa = 6.86PTTTGYY344 pKa = 11.04DD345 pKa = 3.13SDD347 pKa = 4.85FNFGNLALSMAGDD360 pKa = 3.84FTDD363 pKa = 5.53LLFTLSGTKK372 pKa = 7.99ITKK375 pKa = 9.89EE376 pKa = 3.91IVPMAFDD383 pKa = 3.31VLKK386 pKa = 10.9AVLNDD391 pKa = 3.23EE392 pKa = 4.41SGNYY396 pKa = 7.0QTVEE400 pKa = 4.13DD401 pKa = 4.18ATAAHH406 pKa = 6.72YY407 pKa = 7.89ATITDD412 pKa = 4.22FNPRR416 pKa = 11.84EE417 pKa = 4.08DD418 pKa = 3.75VFIVPLASTGNINIDD433 pKa = 2.98ITTDD437 pKa = 4.19GIGNSAFALDD447 pKa = 4.01YY448 pKa = 11.48VNGNGRR454 pKa = 11.84LAKK457 pKa = 10.15VYY459 pKa = 9.61YY460 pKa = 9.95DD461 pKa = 3.74SSISDD466 pKa = 3.6SAQNFYY472 pKa = 11.1GKK474 pKa = 10.58ALLDD478 pKa = 3.39SALIIEE484 pKa = 4.82NDD486 pKa = 3.1AVTYY490 pKa = 10.74GGDD493 pKa = 3.43DD494 pKa = 3.34SGSIFSSSDD503 pKa = 3.05LYY505 pKa = 11.64ADD507 pKa = 4.73LSDD510 pKa = 4.84DD511 pKa = 3.91VKK513 pKa = 11.47ASLGNLSGRR522 pKa = 11.84YY523 pKa = 8.32MILGAFSGSYY533 pKa = 10.61YY534 pKa = 10.55EE535 pKa = 5.2DD536 pKa = 4.49LYY538 pKa = 11.6GNSDD542 pKa = 3.21EE543 pKa = 5.85GMYY546 pKa = 8.33GTNYY550 pKa = 10.51GDD552 pKa = 3.65VLYY555 pKa = 10.09GYY557 pKa = 9.92QKK559 pKa = 9.42EE560 pKa = 4.81TTDD563 pKa = 2.87SWSPQNDD570 pKa = 3.55GNDD573 pKa = 3.2YY574 pKa = 10.47FYY576 pKa = 10.81GYY578 pKa = 9.17TGDD581 pKa = 3.68DD582 pKa = 3.18VFYY585 pKa = 10.95GGGGINYY592 pKa = 8.53FFGGEE597 pKa = 3.84SAYY600 pKa = 11.29NNSGYY605 pKa = 10.54NEE607 pKa = 4.18SADD610 pKa = 3.65NGSDD614 pKa = 3.19TVSYY618 pKa = 11.15EE619 pKa = 4.12DD620 pKa = 3.96ANSGAGVNIDD630 pKa = 3.98LSVTATDD637 pKa = 3.53QNGTYY642 pKa = 9.89TLVSANGFISDD653 pKa = 3.83QNVEE657 pKa = 4.26GVDD660 pKa = 3.11RR661 pKa = 11.84LYY663 pKa = 10.98SIEE666 pKa = 4.5NIIGTEE672 pKa = 3.86FADD675 pKa = 4.44TITGNSGDD683 pKa = 3.87NVLGGSGGNDD693 pKa = 2.74ILKK696 pKa = 10.66GEE698 pKa = 4.23SGNDD702 pKa = 2.76IFYY705 pKa = 10.91GGSGSNEE712 pKa = 3.17IWGGSGTDD720 pKa = 3.08TVDD723 pKa = 3.29YY724 pKa = 10.84SRR726 pKa = 11.84STSGVSGSTVNWSKK740 pKa = 9.22ITHH743 pKa = 5.89SEE745 pKa = 4.85GIDD748 pKa = 3.22QFKK751 pKa = 11.2NNDD754 pKa = 3.35GGGVEE759 pKa = 4.32IVIGSDD765 pKa = 3.27YY766 pKa = 11.32DD767 pKa = 3.74DD768 pKa = 5.17TITGHH773 pKa = 5.69WVANTLYY780 pKa = 10.51GGAGNDD786 pKa = 3.26ILNGGGGNYY795 pKa = 10.3ADD797 pKa = 4.35TLDD800 pKa = 4.49GGSGSDD806 pKa = 3.39TLRR809 pKa = 11.84GANGNDD815 pKa = 3.35TLIGGGGQDD824 pKa = 2.82RR825 pKa = 11.84FEE827 pKa = 4.74FDD829 pKa = 3.97SDD831 pKa = 4.38DD832 pKa = 3.87GTDD835 pKa = 3.35TITDD839 pKa = 3.55YY840 pKa = 11.55SSNSDD845 pKa = 3.45TLSFDD850 pKa = 3.16GDD852 pKa = 4.16VVVTSYY858 pKa = 11.81SDD860 pKa = 3.78GSDD863 pKa = 3.43TYY865 pKa = 11.64VSAGDD870 pKa = 3.63TTVKK874 pKa = 10.58LVGVSNGDD882 pKa = 3.21IASGFSYY889 pKa = 10.82QSDD892 pKa = 3.79TNTTEE897 pKa = 4.11NGPSGNTVICTYY909 pKa = 7.25MHH911 pKa = 7.38EE912 pKa = 4.22IGEE915 pKa = 4.35ISDD918 pKa = 5.31EE919 pKa = 4.34VYY921 pKa = 10.52QWDD924 pKa = 3.55QYY926 pKa = 10.94YY927 pKa = 10.89GRR929 pKa = 11.84EE930 pKa = 4.08FLGKK934 pKa = 7.66TVLRR938 pKa = 11.84GYY940 pKa = 9.9HH941 pKa = 5.18YY942 pKa = 9.85WAIPFVRR949 pKa = 11.84AMRR952 pKa = 11.84RR953 pKa = 11.84NPRR956 pKa = 11.84LTQVMIPLAKK966 pKa = 10.33AWTQEE971 pKa = 3.8MAHH974 pKa = 6.55RR975 pKa = 11.84CDD977 pKa = 4.19PDD979 pKa = 3.3NHH981 pKa = 5.06EE982 pKa = 4.41HH983 pKa = 6.44NRR985 pKa = 11.84LGSLALAVGVPPSRR999 pKa = 11.84IIGSVLEE1006 pKa = 3.98ANYY1009 pKa = 10.32RR1010 pKa = 11.84RR1011 pKa = 11.84LSALRR1016 pKa = 11.84KK1017 pKa = 9.64SAVIDD1022 pKa = 3.59SS1023 pKa = 3.93
MM1 pKa = 7.55RR2 pKa = 11.84PSRR5 pKa = 11.84SLLVDD10 pKa = 5.0PISGPSHH17 pKa = 6.29GKK19 pKa = 10.09ALLSDD24 pKa = 3.68TLLRR28 pKa = 11.84AQNALQDD35 pKa = 4.21FVAKK39 pKa = 10.48DD40 pKa = 3.54VFDD43 pKa = 3.82TKK45 pKa = 11.07VFTVAFGEE53 pKa = 4.23NYY55 pKa = 10.17DD56 pKa = 4.14AIIFEE61 pKa = 4.45SLIDD65 pKa = 3.43QYY67 pKa = 11.95RR68 pKa = 11.84NGNFSSLPSIEE79 pKa = 4.27LRR81 pKa = 11.84SGLEE85 pKa = 3.73LQGADD90 pKa = 3.15GVYY93 pKa = 10.09VEE95 pKa = 4.85SLNAIFISDD104 pKa = 3.91TFYY107 pKa = 11.88GDD109 pKa = 4.88ANDD112 pKa = 4.07TQQVAVVLEE121 pKa = 4.64EE122 pKa = 3.9IGHH125 pKa = 7.02AIDD128 pKa = 4.74ALINKK133 pKa = 9.21SDD135 pKa = 3.61SKK137 pKa = 11.22GDD139 pKa = 3.42EE140 pKa = 3.59GRR142 pKa = 11.84IFAKK146 pKa = 10.45LVLGQEE152 pKa = 4.14MSDD155 pKa = 3.33EE156 pKa = 4.57VVAEE160 pKa = 3.99LHH162 pKa = 7.16AEE164 pKa = 4.17DD165 pKa = 5.9DD166 pKa = 4.31SLTLTVDD173 pKa = 3.51GQTVLAEE180 pKa = 4.35ASTKK184 pKa = 10.18VWDD187 pKa = 4.01SGDD190 pKa = 3.01IGGPYY195 pKa = 8.44ATTWKK200 pKa = 10.48YY201 pKa = 10.24ILKK204 pKa = 9.39TIDD207 pKa = 3.31TPYY210 pKa = 11.17SMSSTYY216 pKa = 10.37FEE218 pKa = 4.21NTLKK222 pKa = 10.44HH223 pKa = 6.19DD224 pKa = 4.06RR225 pKa = 11.84TWSDD229 pKa = 3.57FIGNKK234 pKa = 6.94TKK236 pKa = 10.29YY237 pKa = 8.24YY238 pKa = 10.23EE239 pKa = 4.18YY240 pKa = 10.35YY241 pKa = 10.97GEE243 pKa = 4.38VNGTSGDD250 pKa = 4.09DD251 pKa = 3.73IIIGATDD258 pKa = 3.53TKK260 pKa = 11.0KK261 pKa = 10.44KK262 pKa = 10.07WEE264 pKa = 4.91DD265 pKa = 3.58GNNDD269 pKa = 2.43IYY271 pKa = 11.13TWLRR275 pKa = 11.84AQEE278 pKa = 3.88EE279 pKa = 4.9TINGGSGDD287 pKa = 4.24DD288 pKa = 4.42YY289 pKa = 11.46IDD291 pKa = 4.41GGDD294 pKa = 4.08GSDD297 pKa = 4.32TINGGAGNDD306 pKa = 4.09LIFTGDD312 pKa = 3.85LSDD315 pKa = 5.17GSDD318 pKa = 3.7DD319 pKa = 4.69DD320 pKa = 5.4VITGGSGADD329 pKa = 3.2TFYY332 pKa = 10.74TGIITHH338 pKa = 6.86PTTTGYY344 pKa = 11.04DD345 pKa = 3.13SDD347 pKa = 4.85FNFGNLALSMAGDD360 pKa = 3.84FTDD363 pKa = 5.53LLFTLSGTKK372 pKa = 7.99ITKK375 pKa = 9.89EE376 pKa = 3.91IVPMAFDD383 pKa = 3.31VLKK386 pKa = 10.9AVLNDD391 pKa = 3.23EE392 pKa = 4.41SGNYY396 pKa = 7.0QTVEE400 pKa = 4.13DD401 pKa = 4.18ATAAHH406 pKa = 6.72YY407 pKa = 7.89ATITDD412 pKa = 4.22FNPRR416 pKa = 11.84EE417 pKa = 4.08DD418 pKa = 3.75VFIVPLASTGNINIDD433 pKa = 2.98ITTDD437 pKa = 4.19GIGNSAFALDD447 pKa = 4.01YY448 pKa = 11.48VNGNGRR454 pKa = 11.84LAKK457 pKa = 10.15VYY459 pKa = 9.61YY460 pKa = 9.95DD461 pKa = 3.74SSISDD466 pKa = 3.6SAQNFYY472 pKa = 11.1GKK474 pKa = 10.58ALLDD478 pKa = 3.39SALIIEE484 pKa = 4.82NDD486 pKa = 3.1AVTYY490 pKa = 10.74GGDD493 pKa = 3.43DD494 pKa = 3.34SGSIFSSSDD503 pKa = 3.05LYY505 pKa = 11.64ADD507 pKa = 4.73LSDD510 pKa = 4.84DD511 pKa = 3.91VKK513 pKa = 11.47ASLGNLSGRR522 pKa = 11.84YY523 pKa = 8.32MILGAFSGSYY533 pKa = 10.61YY534 pKa = 10.55EE535 pKa = 5.2DD536 pKa = 4.49LYY538 pKa = 11.6GNSDD542 pKa = 3.21EE543 pKa = 5.85GMYY546 pKa = 8.33GTNYY550 pKa = 10.51GDD552 pKa = 3.65VLYY555 pKa = 10.09GYY557 pKa = 9.92QKK559 pKa = 9.42EE560 pKa = 4.81TTDD563 pKa = 2.87SWSPQNDD570 pKa = 3.55GNDD573 pKa = 3.2YY574 pKa = 10.47FYY576 pKa = 10.81GYY578 pKa = 9.17TGDD581 pKa = 3.68DD582 pKa = 3.18VFYY585 pKa = 10.95GGGGINYY592 pKa = 8.53FFGGEE597 pKa = 3.84SAYY600 pKa = 11.29NNSGYY605 pKa = 10.54NEE607 pKa = 4.18SADD610 pKa = 3.65NGSDD614 pKa = 3.19TVSYY618 pKa = 11.15EE619 pKa = 4.12DD620 pKa = 3.96ANSGAGVNIDD630 pKa = 3.98LSVTATDD637 pKa = 3.53QNGTYY642 pKa = 9.89TLVSANGFISDD653 pKa = 3.83QNVEE657 pKa = 4.26GVDD660 pKa = 3.11RR661 pKa = 11.84LYY663 pKa = 10.98SIEE666 pKa = 4.5NIIGTEE672 pKa = 3.86FADD675 pKa = 4.44TITGNSGDD683 pKa = 3.87NVLGGSGGNDD693 pKa = 2.74ILKK696 pKa = 10.66GEE698 pKa = 4.23SGNDD702 pKa = 2.76IFYY705 pKa = 10.91GGSGSNEE712 pKa = 3.17IWGGSGTDD720 pKa = 3.08TVDD723 pKa = 3.29YY724 pKa = 10.84SRR726 pKa = 11.84STSGVSGSTVNWSKK740 pKa = 9.22ITHH743 pKa = 5.89SEE745 pKa = 4.85GIDD748 pKa = 3.22QFKK751 pKa = 11.2NNDD754 pKa = 3.35GGGVEE759 pKa = 4.32IVIGSDD765 pKa = 3.27YY766 pKa = 11.32DD767 pKa = 3.74DD768 pKa = 5.17TITGHH773 pKa = 5.69WVANTLYY780 pKa = 10.51GGAGNDD786 pKa = 3.26ILNGGGGNYY795 pKa = 10.3ADD797 pKa = 4.35TLDD800 pKa = 4.49GGSGSDD806 pKa = 3.39TLRR809 pKa = 11.84GANGNDD815 pKa = 3.35TLIGGGGQDD824 pKa = 2.82RR825 pKa = 11.84FEE827 pKa = 4.74FDD829 pKa = 3.97SDD831 pKa = 4.38DD832 pKa = 3.87GTDD835 pKa = 3.35TITDD839 pKa = 3.55YY840 pKa = 11.55SSNSDD845 pKa = 3.45TLSFDD850 pKa = 3.16GDD852 pKa = 4.16VVVTSYY858 pKa = 11.81SDD860 pKa = 3.78GSDD863 pKa = 3.43TYY865 pKa = 11.64VSAGDD870 pKa = 3.63TTVKK874 pKa = 10.58LVGVSNGDD882 pKa = 3.21IASGFSYY889 pKa = 10.82QSDD892 pKa = 3.79TNTTEE897 pKa = 4.11NGPSGNTVICTYY909 pKa = 7.25MHH911 pKa = 7.38EE912 pKa = 4.22IGEE915 pKa = 4.35ISDD918 pKa = 5.31EE919 pKa = 4.34VYY921 pKa = 10.52QWDD924 pKa = 3.55QYY926 pKa = 10.94YY927 pKa = 10.89GRR929 pKa = 11.84EE930 pKa = 4.08FLGKK934 pKa = 7.66TVLRR938 pKa = 11.84GYY940 pKa = 9.9HH941 pKa = 5.18YY942 pKa = 9.85WAIPFVRR949 pKa = 11.84AMRR952 pKa = 11.84RR953 pKa = 11.84NPRR956 pKa = 11.84LTQVMIPLAKK966 pKa = 10.33AWTQEE971 pKa = 3.8MAHH974 pKa = 6.55RR975 pKa = 11.84CDD977 pKa = 4.19PDD979 pKa = 3.3NHH981 pKa = 5.06EE982 pKa = 4.41HH983 pKa = 6.44NRR985 pKa = 11.84LGSLALAVGVPPSRR999 pKa = 11.84IIGSVLEE1006 pKa = 3.98ANYY1009 pKa = 10.32RR1010 pKa = 11.84RR1011 pKa = 11.84LSALRR1016 pKa = 11.84KK1017 pKa = 9.64SAVIDD1022 pKa = 3.59SS1023 pKa = 3.93
Molecular weight: 109.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y0IGN0|A0A1Y0IGN0_9GAMM Paraquat-inducible protein B OS=Oleiphilus messinensis OX=141451 GN=OLMES_5299 PE=4 SV=1
MM1 pKa = 7.94AEE3 pKa = 4.12FLSIGAAAFLLGVAVSTLRR22 pKa = 11.84CWEE25 pKa = 3.88KK26 pKa = 10.5EE27 pKa = 3.8SRR29 pKa = 11.84FFSDD33 pKa = 3.57FRR35 pKa = 11.84TPGGHH40 pKa = 6.3RR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 10.62ALDD46 pKa = 4.28KK47 pKa = 10.97LLAFCGQSTANEE59 pKa = 3.89QRR61 pKa = 11.84RR62 pKa = 11.84TICYY66 pKa = 10.24ARR68 pKa = 11.84VSSHH72 pKa = 6.2DD73 pKa = 3.59QKK75 pKa = 11.66KK76 pKa = 10.55DD77 pKa = 3.21LQTQIARR84 pKa = 11.84LYY86 pKa = 9.97GSRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84KK92 pKa = 7.75NQRR95 pKa = 11.84AIAA98 pKa = 3.94
MM1 pKa = 7.94AEE3 pKa = 4.12FLSIGAAAFLLGVAVSTLRR22 pKa = 11.84CWEE25 pKa = 3.88KK26 pKa = 10.5EE27 pKa = 3.8SRR29 pKa = 11.84FFSDD33 pKa = 3.57FRR35 pKa = 11.84TPGGHH40 pKa = 6.3RR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 10.62ALDD46 pKa = 4.28KK47 pKa = 10.97LLAFCGQSTANEE59 pKa = 3.89QRR61 pKa = 11.84RR62 pKa = 11.84TICYY66 pKa = 10.24ARR68 pKa = 11.84VSSHH72 pKa = 6.2DD73 pKa = 3.59QKK75 pKa = 11.66KK76 pKa = 10.55DD77 pKa = 3.21LQTQIARR84 pKa = 11.84LYY86 pKa = 9.97GSRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84KK92 pKa = 7.75NQRR95 pKa = 11.84AIAA98 pKa = 3.94
Molecular weight: 11.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1829888 |
19 |
7866 |
335.4 |
37.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.581 ± 0.035 | 1.043 ± 0.012 |
5.663 ± 0.031 | 6.396 ± 0.033 |
4.108 ± 0.023 | 6.811 ± 0.04 |
2.305 ± 0.018 | 6.169 ± 0.024 |
4.736 ± 0.032 | 10.519 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.314 ± 0.018 | 4.087 ± 0.024 |
4.238 ± 0.023 | 4.609 ± 0.023 |
5.284 ± 0.026 | 6.734 ± 0.028 |
5.382 ± 0.03 | 6.795 ± 0.032 |
1.253 ± 0.013 | 2.974 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
