
Merida-like virus KE-2017a
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
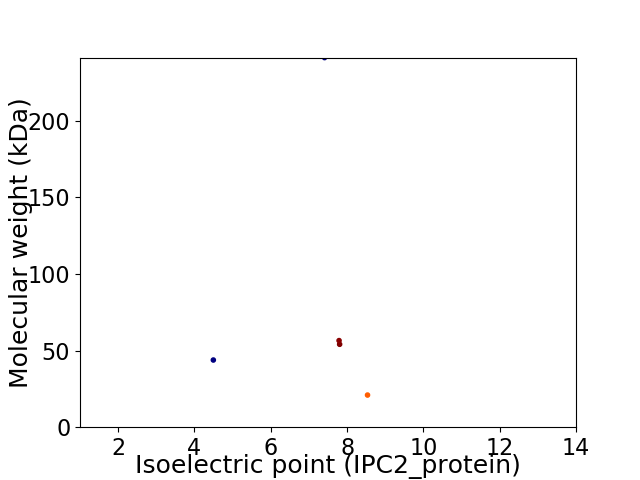
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U9YAZ4|A0A1U9YAZ4_9RHAB GDP polyribonucleotidyltransferase OS=Merida-like virus KE-2017a OX=1969830 PE=4 SV=1
MM1 pKa = 7.61SEE3 pKa = 4.06PGGEE7 pKa = 4.16SVQTTKK13 pKa = 10.7SEE15 pKa = 4.03EE16 pKa = 4.07EE17 pKa = 3.53IRR19 pKa = 11.84AEE21 pKa = 3.9SFYY24 pKa = 11.35SVGSDD29 pKa = 3.56LPQEE33 pKa = 4.43SVSCLSALLEE43 pKa = 3.99YY44 pKa = 10.67DD45 pKa = 3.67QEE47 pKa = 4.41GLEE50 pKa = 4.33KK51 pKa = 10.2TFSQIDD57 pKa = 3.85LEE59 pKa = 4.82DD60 pKa = 4.38PLTGEE65 pKa = 4.46GGGGSVQVAVPWGKK79 pKa = 9.77IPEE82 pKa = 4.27EE83 pKa = 4.06KK84 pKa = 9.6QAEE87 pKa = 4.27YY88 pKa = 11.06LKK90 pKa = 11.01LLSSYY95 pKa = 10.26NIPPAEE101 pKa = 4.2TSDD104 pKa = 3.75EE105 pKa = 4.13EE106 pKa = 5.44SGVDD110 pKa = 4.75DD111 pKa = 5.37DD112 pKa = 4.88KK113 pKa = 11.7QSSGTPSLPPTSPSLPSEE131 pKa = 4.1RR132 pKa = 11.84GNRR135 pKa = 11.84PLSPFLFSDD144 pKa = 4.03EE145 pKa = 4.67EE146 pKa = 4.11YY147 pKa = 10.94SEE149 pKa = 4.37YY150 pKa = 11.31VDD152 pKa = 3.36IFVPEE157 pKa = 4.07VLQDD161 pKa = 3.39VSLRR165 pKa = 11.84TPLMSLLRR173 pKa = 11.84DD174 pKa = 3.7VVRR177 pKa = 11.84VMVNSKK183 pKa = 10.66NEE185 pKa = 4.05PLYY188 pKa = 10.98SLQKK192 pKa = 9.19YY193 pKa = 8.31DD194 pKa = 3.75LRR196 pKa = 11.84EE197 pKa = 3.99GTVRR201 pKa = 11.84LARR204 pKa = 11.84VGHH207 pKa = 7.06LFTYY211 pKa = 10.17CPEE214 pKa = 4.21GTPIPVLEE222 pKa = 4.27QRR224 pKa = 11.84GSPKK228 pKa = 9.61PQKK231 pKa = 10.28KK232 pKa = 10.48GIFATTQKK240 pKa = 11.14DD241 pKa = 3.25LTAPSVRR248 pKa = 11.84EE249 pKa = 3.89TTSRR253 pKa = 11.84GVTADD258 pKa = 5.14PIPSGSKK265 pKa = 8.79TLADD269 pKa = 3.69SASGQINQVSQTFEE283 pKa = 4.19EE284 pKa = 4.64TWPSLEE290 pKa = 4.66HH291 pKa = 6.65SGSPLNLEE299 pKa = 3.97RR300 pKa = 11.84GVNVQRR306 pKa = 11.84AVSPSSSSHH315 pKa = 5.3YY316 pKa = 10.24SQTSSVEE323 pKa = 4.27SNSDD327 pKa = 3.32NEE329 pKa = 4.14EE330 pKa = 4.17EE331 pKa = 4.21IEE333 pKa = 4.5VMQEE337 pKa = 4.17TYY339 pKa = 11.46SFMCPIYY346 pKa = 10.19GSSEE350 pKa = 3.75MSVKK354 pKa = 10.01VFPGYY359 pKa = 9.73EE360 pKa = 3.95VEE362 pKa = 4.63PLLKK366 pKa = 10.64LGLCPPEE373 pKa = 3.86IFRR376 pKa = 11.84SILRR380 pKa = 11.84DD381 pKa = 3.37RR382 pKa = 11.84GLLRR386 pKa = 11.84ALEE389 pKa = 4.2AVLDD393 pKa = 3.7IGQTHH398 pKa = 5.3MTT400 pKa = 3.99
MM1 pKa = 7.61SEE3 pKa = 4.06PGGEE7 pKa = 4.16SVQTTKK13 pKa = 10.7SEE15 pKa = 4.03EE16 pKa = 4.07EE17 pKa = 3.53IRR19 pKa = 11.84AEE21 pKa = 3.9SFYY24 pKa = 11.35SVGSDD29 pKa = 3.56LPQEE33 pKa = 4.43SVSCLSALLEE43 pKa = 3.99YY44 pKa = 10.67DD45 pKa = 3.67QEE47 pKa = 4.41GLEE50 pKa = 4.33KK51 pKa = 10.2TFSQIDD57 pKa = 3.85LEE59 pKa = 4.82DD60 pKa = 4.38PLTGEE65 pKa = 4.46GGGGSVQVAVPWGKK79 pKa = 9.77IPEE82 pKa = 4.27EE83 pKa = 4.06KK84 pKa = 9.6QAEE87 pKa = 4.27YY88 pKa = 11.06LKK90 pKa = 11.01LLSSYY95 pKa = 10.26NIPPAEE101 pKa = 4.2TSDD104 pKa = 3.75EE105 pKa = 4.13EE106 pKa = 5.44SGVDD110 pKa = 4.75DD111 pKa = 5.37DD112 pKa = 4.88KK113 pKa = 11.7QSSGTPSLPPTSPSLPSEE131 pKa = 4.1RR132 pKa = 11.84GNRR135 pKa = 11.84PLSPFLFSDD144 pKa = 4.03EE145 pKa = 4.67EE146 pKa = 4.11YY147 pKa = 10.94SEE149 pKa = 4.37YY150 pKa = 11.31VDD152 pKa = 3.36IFVPEE157 pKa = 4.07VLQDD161 pKa = 3.39VSLRR165 pKa = 11.84TPLMSLLRR173 pKa = 11.84DD174 pKa = 3.7VVRR177 pKa = 11.84VMVNSKK183 pKa = 10.66NEE185 pKa = 4.05PLYY188 pKa = 10.98SLQKK192 pKa = 9.19YY193 pKa = 8.31DD194 pKa = 3.75LRR196 pKa = 11.84EE197 pKa = 3.99GTVRR201 pKa = 11.84LARR204 pKa = 11.84VGHH207 pKa = 7.06LFTYY211 pKa = 10.17CPEE214 pKa = 4.21GTPIPVLEE222 pKa = 4.27QRR224 pKa = 11.84GSPKK228 pKa = 9.61PQKK231 pKa = 10.28KK232 pKa = 10.48GIFATTQKK240 pKa = 11.14DD241 pKa = 3.25LTAPSVRR248 pKa = 11.84EE249 pKa = 3.89TTSRR253 pKa = 11.84GVTADD258 pKa = 5.14PIPSGSKK265 pKa = 8.79TLADD269 pKa = 3.69SASGQINQVSQTFEE283 pKa = 4.19EE284 pKa = 4.64TWPSLEE290 pKa = 4.66HH291 pKa = 6.65SGSPLNLEE299 pKa = 3.97RR300 pKa = 11.84GVNVQRR306 pKa = 11.84AVSPSSSSHH315 pKa = 5.3YY316 pKa = 10.24SQTSSVEE323 pKa = 4.27SNSDD327 pKa = 3.32NEE329 pKa = 4.14EE330 pKa = 4.17EE331 pKa = 4.21IEE333 pKa = 4.5VMQEE337 pKa = 4.17TYY339 pKa = 11.46SFMCPIYY346 pKa = 10.19GSSEE350 pKa = 3.75MSVKK354 pKa = 10.01VFPGYY359 pKa = 9.73EE360 pKa = 3.95VEE362 pKa = 4.63PLLKK366 pKa = 10.64LGLCPPEE373 pKa = 3.86IFRR376 pKa = 11.84SILRR380 pKa = 11.84DD381 pKa = 3.37RR382 pKa = 11.84GLLRR386 pKa = 11.84ALEE389 pKa = 4.2AVLDD393 pKa = 3.7IGQTHH398 pKa = 5.3MTT400 pKa = 3.99
Molecular weight: 43.84 kDa
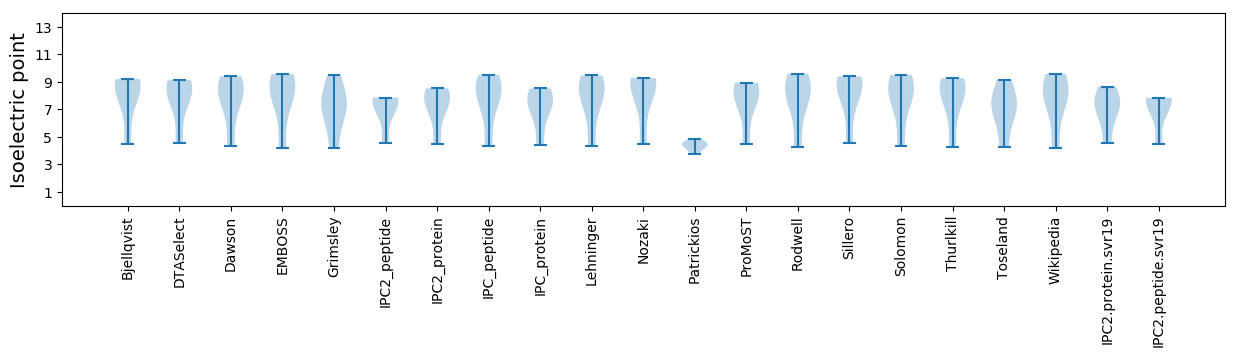
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U9YAZ7|A0A1U9YAZ7_9RHAB Nucleocapsid protein OS=Merida-like virus KE-2017a OX=1969830 PE=4 SV=1
MM1 pKa = 7.41SFVSMLKK8 pKa = 10.52GKK10 pKa = 10.19SSPSKK15 pKa = 9.67RR16 pKa = 11.84SPSTGHH22 pKa = 6.3VPFQGRR28 pKa = 11.84LDD30 pKa = 3.51MSLTIFTDD38 pKa = 3.62TPYY41 pKa = 11.48SSMEE45 pKa = 3.83EE46 pKa = 4.0AFLHH50 pKa = 6.56LIEE53 pKa = 4.43IQDD56 pKa = 4.29YY57 pKa = 10.47ISEE60 pKa = 4.18TRR62 pKa = 11.84DD63 pKa = 3.34LKK65 pKa = 11.16GLYY68 pKa = 9.7LGLIAVSILFSKK80 pKa = 10.75CDD82 pKa = 3.46ARR84 pKa = 11.84GSGVMYY90 pKa = 10.82TMDD93 pKa = 4.41LKK95 pKa = 11.33DD96 pKa = 4.11LVSLISTDD104 pKa = 3.13SALASGPLRR113 pKa = 11.84TFSYY117 pKa = 10.92EE118 pKa = 3.7RR119 pKa = 11.84THH121 pKa = 7.18DD122 pKa = 3.89LRR124 pKa = 11.84GKK126 pKa = 7.42TALVRR131 pKa = 11.84FRR133 pKa = 11.84LDD135 pKa = 3.61FEE137 pKa = 4.5QSGLDD142 pKa = 3.82CEE144 pKa = 4.79SLLSVLMKK152 pKa = 10.38RR153 pKa = 11.84NKK155 pKa = 9.87NKK157 pKa = 10.43EE158 pKa = 4.14VMSSSTSYY166 pKa = 11.12LKK168 pKa = 10.58LWRR171 pKa = 11.84LKK173 pKa = 10.39MIDD176 pKa = 3.84YY177 pKa = 10.71KK178 pKa = 10.92RR179 pKa = 11.84GTLFSLSS186 pKa = 3.5
MM1 pKa = 7.41SFVSMLKK8 pKa = 10.52GKK10 pKa = 10.19SSPSKK15 pKa = 9.67RR16 pKa = 11.84SPSTGHH22 pKa = 6.3VPFQGRR28 pKa = 11.84LDD30 pKa = 3.51MSLTIFTDD38 pKa = 3.62TPYY41 pKa = 11.48SSMEE45 pKa = 3.83EE46 pKa = 4.0AFLHH50 pKa = 6.56LIEE53 pKa = 4.43IQDD56 pKa = 4.29YY57 pKa = 10.47ISEE60 pKa = 4.18TRR62 pKa = 11.84DD63 pKa = 3.34LKK65 pKa = 11.16GLYY68 pKa = 9.7LGLIAVSILFSKK80 pKa = 10.75CDD82 pKa = 3.46ARR84 pKa = 11.84GSGVMYY90 pKa = 10.82TMDD93 pKa = 4.41LKK95 pKa = 11.33DD96 pKa = 4.11LVSLISTDD104 pKa = 3.13SALASGPLRR113 pKa = 11.84TFSYY117 pKa = 10.92EE118 pKa = 3.7RR119 pKa = 11.84THH121 pKa = 7.18DD122 pKa = 3.89LRR124 pKa = 11.84GKK126 pKa = 7.42TALVRR131 pKa = 11.84FRR133 pKa = 11.84LDD135 pKa = 3.61FEE137 pKa = 4.5QSGLDD142 pKa = 3.82CEE144 pKa = 4.79SLLSVLMKK152 pKa = 10.38RR153 pKa = 11.84NKK155 pKa = 9.87NKK157 pKa = 10.43EE158 pKa = 4.14VMSSSTSYY166 pKa = 11.12LKK168 pKa = 10.58LWRR171 pKa = 11.84LKK173 pKa = 10.39MIDD176 pKa = 3.84YY177 pKa = 10.71KK178 pKa = 10.92RR179 pKa = 11.84GTLFSLSS186 pKa = 3.5
Molecular weight: 20.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3708 |
186 |
2136 |
741.6 |
83.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.529 ± 0.373 | 1.618 ± 0.243 |
5.583 ± 0.371 | 6.041 ± 0.795 |
4.477 ± 0.593 | 7.012 ± 0.073 |
2.05 ± 0.231 | 5.987 ± 0.587 |
5.717 ± 0.537 | 10.14 ± 0.401 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.238 ± 0.283 | 3.155 ± 0.358 |
5.097 ± 0.622 | 3.371 ± 0.232 |
5.906 ± 0.486 | 9.709 ± 0.83 |
5.609 ± 0.264 | 5.636 ± 0.416 |
1.888 ± 0.292 | 3.236 ± 0.177 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
