
Heterobasidion partitivirus 15
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
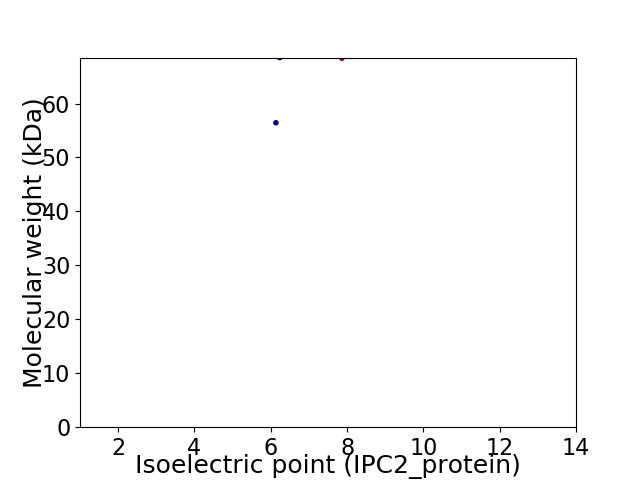
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
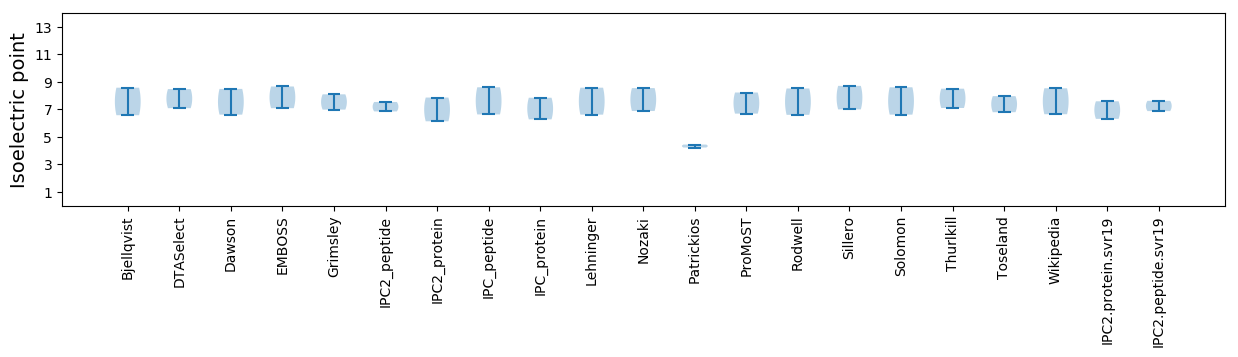
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8PR84|W8PR84_9VIRU RNA-dependent RNA polymerase OS=Heterobasidion partitivirus 15 OX=1469908 PE=4 SV=1
MM1 pKa = 7.55SATKK5 pKa = 10.05MEE7 pKa = 5.36DD8 pKa = 3.52YY9 pKa = 11.49LNGKK13 pKa = 9.09LDD15 pKa = 3.62AAALNKK21 pKa = 10.2ADD23 pKa = 3.46IEE25 pKa = 4.42AYY27 pKa = 10.09AAAQIKK33 pKa = 8.77AQKK36 pKa = 10.45DD37 pKa = 3.56LIEE40 pKa = 4.94ASTSASKK47 pKa = 10.87DD48 pKa = 3.25AGSDD52 pKa = 3.5PLGMRR57 pKa = 11.84DD58 pKa = 3.39VHH60 pKa = 6.88KK61 pKa = 10.43QAQDD65 pKa = 3.22SVQAQSQPTSTNLRR79 pKa = 11.84PPRR82 pKa = 11.84FQKK85 pKa = 10.46VDD87 pKa = 3.3DD88 pKa = 3.84TDD90 pKa = 5.0AINAHH95 pKa = 6.58RR96 pKa = 11.84LVLMDD101 pKa = 5.5DD102 pKa = 4.28PFTHH106 pKa = 6.39VSKK109 pKa = 10.8FGNNFVVPDD118 pKa = 4.07FHH120 pKa = 7.85HH121 pKa = 7.11AFTMLDD127 pKa = 3.27EE128 pKa = 4.91MDD130 pKa = 5.24NIMLSTKK137 pKa = 10.15KK138 pKa = 9.39FTDD141 pKa = 3.45ISEE144 pKa = 4.1TWTPLMSRR152 pKa = 11.84YY153 pKa = 9.61FFGSLLLVHH162 pKa = 6.43TLRR165 pKa = 11.84VQRR168 pKa = 11.84EE169 pKa = 3.9ADD171 pKa = 3.24AVTPTVANFLDD182 pKa = 3.88QFEE185 pKa = 5.02KK186 pKa = 10.89DD187 pKa = 4.12FPLHH191 pKa = 6.64SIPIPGPMVNFFEE204 pKa = 5.07CLAASNVAVAGFDD217 pKa = 3.53PVSPILPKK225 pKa = 10.19KK226 pKa = 10.75ANVTTRR232 pKa = 11.84AKK234 pKa = 10.92LLFNNNLAGRR244 pKa = 11.84IPHH247 pKa = 6.54PLIMLDD253 pKa = 3.54QMHH256 pKa = 6.13VLINAMGNGDD266 pKa = 3.79RR267 pKa = 11.84ATRR270 pKa = 11.84ISNVANCSVFFNGIFSIGMSNAASADD296 pKa = 3.85PYY298 pKa = 10.74EE299 pKa = 4.34QTPVHH304 pKa = 6.27AVNTANHH311 pKa = 5.99IAWVFSYY318 pKa = 10.76PDD320 pKa = 2.9VAYY323 pKa = 9.29PVLSSYY329 pKa = 10.77NAASSFVDD337 pKa = 4.15NINQLRR343 pKa = 11.84RR344 pKa = 11.84VLPARR349 pKa = 11.84LDD351 pKa = 3.45LSGAGNATQLEE362 pKa = 4.53WPEE365 pKa = 3.94FVSIMEE371 pKa = 3.97YY372 pKa = 10.19PRR374 pKa = 11.84LFRR377 pKa = 11.84YY378 pKa = 9.6ILRR381 pKa = 11.84LMAEE385 pKa = 4.11YY386 pKa = 11.02SKK388 pKa = 10.57FWKK391 pKa = 10.32GSKK394 pKa = 10.27SLDD397 pKa = 4.14LIPLAGKK404 pKa = 9.66PSLQVAFTKK413 pKa = 10.7DD414 pKa = 3.69GAEE417 pKa = 3.81HH418 pKa = 7.34GYY420 pKa = 7.58PTTRR424 pKa = 11.84FPTFTHH430 pKa = 6.5SFTGAVRR437 pKa = 11.84RR438 pKa = 11.84SQISPADD445 pKa = 4.69RR446 pKa = 11.84IDD448 pKa = 3.62AATSVLNYY456 pKa = 10.58SHH458 pKa = 7.18LADD461 pKa = 4.17GGGIDD466 pKa = 4.28RR467 pKa = 11.84ANPGIPRR474 pKa = 11.84QGGPFFDD481 pKa = 5.0DD482 pKa = 3.62SEE484 pKa = 4.5IVLRR488 pKa = 11.84TRR490 pKa = 11.84NINPSLGYY498 pKa = 10.73GSVATEE504 pKa = 4.38KK505 pKa = 10.95YY506 pKa = 9.91HH507 pKa = 7.22LKK509 pKa = 8.2NTKK512 pKa = 9.61
MM1 pKa = 7.55SATKK5 pKa = 10.05MEE7 pKa = 5.36DD8 pKa = 3.52YY9 pKa = 11.49LNGKK13 pKa = 9.09LDD15 pKa = 3.62AAALNKK21 pKa = 10.2ADD23 pKa = 3.46IEE25 pKa = 4.42AYY27 pKa = 10.09AAAQIKK33 pKa = 8.77AQKK36 pKa = 10.45DD37 pKa = 3.56LIEE40 pKa = 4.94ASTSASKK47 pKa = 10.87DD48 pKa = 3.25AGSDD52 pKa = 3.5PLGMRR57 pKa = 11.84DD58 pKa = 3.39VHH60 pKa = 6.88KK61 pKa = 10.43QAQDD65 pKa = 3.22SVQAQSQPTSTNLRR79 pKa = 11.84PPRR82 pKa = 11.84FQKK85 pKa = 10.46VDD87 pKa = 3.3DD88 pKa = 3.84TDD90 pKa = 5.0AINAHH95 pKa = 6.58RR96 pKa = 11.84LVLMDD101 pKa = 5.5DD102 pKa = 4.28PFTHH106 pKa = 6.39VSKK109 pKa = 10.8FGNNFVVPDD118 pKa = 4.07FHH120 pKa = 7.85HH121 pKa = 7.11AFTMLDD127 pKa = 3.27EE128 pKa = 4.91MDD130 pKa = 5.24NIMLSTKK137 pKa = 10.15KK138 pKa = 9.39FTDD141 pKa = 3.45ISEE144 pKa = 4.1TWTPLMSRR152 pKa = 11.84YY153 pKa = 9.61FFGSLLLVHH162 pKa = 6.43TLRR165 pKa = 11.84VQRR168 pKa = 11.84EE169 pKa = 3.9ADD171 pKa = 3.24AVTPTVANFLDD182 pKa = 3.88QFEE185 pKa = 5.02KK186 pKa = 10.89DD187 pKa = 4.12FPLHH191 pKa = 6.64SIPIPGPMVNFFEE204 pKa = 5.07CLAASNVAVAGFDD217 pKa = 3.53PVSPILPKK225 pKa = 10.19KK226 pKa = 10.75ANVTTRR232 pKa = 11.84AKK234 pKa = 10.92LLFNNNLAGRR244 pKa = 11.84IPHH247 pKa = 6.54PLIMLDD253 pKa = 3.54QMHH256 pKa = 6.13VLINAMGNGDD266 pKa = 3.79RR267 pKa = 11.84ATRR270 pKa = 11.84ISNVANCSVFFNGIFSIGMSNAASADD296 pKa = 3.85PYY298 pKa = 10.74EE299 pKa = 4.34QTPVHH304 pKa = 6.27AVNTANHH311 pKa = 5.99IAWVFSYY318 pKa = 10.76PDD320 pKa = 2.9VAYY323 pKa = 9.29PVLSSYY329 pKa = 10.77NAASSFVDD337 pKa = 4.15NINQLRR343 pKa = 11.84RR344 pKa = 11.84VLPARR349 pKa = 11.84LDD351 pKa = 3.45LSGAGNATQLEE362 pKa = 4.53WPEE365 pKa = 3.94FVSIMEE371 pKa = 3.97YY372 pKa = 10.19PRR374 pKa = 11.84LFRR377 pKa = 11.84YY378 pKa = 9.6ILRR381 pKa = 11.84LMAEE385 pKa = 4.11YY386 pKa = 11.02SKK388 pKa = 10.57FWKK391 pKa = 10.32GSKK394 pKa = 10.27SLDD397 pKa = 4.14LIPLAGKK404 pKa = 9.66PSLQVAFTKK413 pKa = 10.7DD414 pKa = 3.69GAEE417 pKa = 3.81HH418 pKa = 7.34GYY420 pKa = 7.58PTTRR424 pKa = 11.84FPTFTHH430 pKa = 6.5SFTGAVRR437 pKa = 11.84RR438 pKa = 11.84SQISPADD445 pKa = 4.69RR446 pKa = 11.84IDD448 pKa = 3.62AATSVLNYY456 pKa = 10.58SHH458 pKa = 7.18LADD461 pKa = 4.17GGGIDD466 pKa = 4.28RR467 pKa = 11.84ANPGIPRR474 pKa = 11.84QGGPFFDD481 pKa = 5.0DD482 pKa = 3.62SEE484 pKa = 4.5IVLRR488 pKa = 11.84TRR490 pKa = 11.84NINPSLGYY498 pKa = 10.73GSVATEE504 pKa = 4.38KK505 pKa = 10.95YY506 pKa = 9.91HH507 pKa = 7.22LKK509 pKa = 8.2NTKK512 pKa = 9.61
Molecular weight: 56.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8PR84|W8PR84_9VIRU RNA-dependent RNA polymerase OS=Heterobasidion partitivirus 15 OX=1469908 PE=4 SV=1
MM1 pKa = 7.53FSLLTWLSTKK11 pKa = 10.18FNALFDD17 pKa = 4.19SVYY20 pKa = 9.57STRR23 pKa = 11.84FEE25 pKa = 4.79HH26 pKa = 6.93NFRR29 pKa = 11.84FLYY32 pKa = 10.05KK33 pKa = 10.18AAPLTTPHH41 pKa = 7.19RR42 pKa = 11.84DD43 pKa = 2.99EE44 pKa = 4.19TKK46 pKa = 8.62YY47 pKa = 11.03AEE49 pKa = 3.93YY50 pKa = 10.0QAYY53 pKa = 9.82VKK55 pKa = 9.91STLDD59 pKa = 3.44HH60 pKa = 6.4VLLGPDD66 pKa = 2.8AHH68 pKa = 6.9HH69 pKa = 6.76IVNGYY74 pKa = 7.96HH75 pKa = 6.58HH76 pKa = 7.84PIATLDD82 pKa = 3.51AVINTFKK89 pKa = 11.0KK90 pKa = 10.79GDD92 pKa = 3.96LPDD95 pKa = 3.67HH96 pKa = 7.09KK97 pKa = 10.43IPKK100 pKa = 8.97DD101 pKa = 3.27LHH103 pKa = 6.49YY104 pKa = 11.16YY105 pKa = 9.9SALLEE110 pKa = 4.3TTKK113 pKa = 10.69RR114 pKa = 11.84FAPPQKK120 pKa = 10.07LRR122 pKa = 11.84PVHH125 pKa = 6.3FADD128 pKa = 4.35LRR130 pKa = 11.84FYY132 pKa = 10.37KK133 pKa = 10.12WNWHH137 pKa = 6.26PNVEE141 pKa = 4.17EE142 pKa = 4.25PFRR145 pKa = 11.84SEE147 pKa = 5.57AEE149 pKa = 3.86LQQAVEE155 pKa = 4.24AAYY158 pKa = 9.87HH159 pKa = 6.64AGLLPDD165 pKa = 3.5GRR167 pKa = 11.84MSFGNLKK174 pKa = 9.49NVVFIRR180 pKa = 11.84VRR182 pKa = 11.84EE183 pKa = 3.89WLHH186 pKa = 4.79QIKK189 pKa = 10.09RR190 pKa = 11.84AQITNPNTLYY200 pKa = 10.29PLINIHH206 pKa = 6.06VKK208 pKa = 9.89PALTTPDD215 pKa = 3.12EE216 pKa = 4.25TKK218 pKa = 10.01IRR220 pKa = 11.84VVYY223 pKa = 10.01GVSKK227 pKa = 10.35LHH229 pKa = 5.83VLPEE233 pKa = 4.17AMFFWPLFRR242 pKa = 11.84HH243 pKa = 5.55YY244 pKa = 10.99LDD246 pKa = 3.84NRR248 pKa = 11.84KK249 pKa = 9.37EE250 pKa = 4.43SPLLWGFEE258 pKa = 4.33TILGGMAVLHH268 pKa = 6.28NMMTIPRR275 pKa = 11.84LYY277 pKa = 8.89FQTFVMVDD285 pKa = 2.48WSGFDD290 pKa = 3.43LRR292 pKa = 11.84SLFEE296 pKa = 4.72PIRR299 pKa = 11.84TDD301 pKa = 3.62VFPAWRR307 pKa = 11.84TYY309 pKa = 10.68FDD311 pKa = 4.42FNNGYY316 pKa = 9.64IPTRR320 pKa = 11.84FYY322 pKa = 10.21RR323 pKa = 11.84TSKK326 pKa = 10.95ADD328 pKa = 3.52PEE330 pKa = 4.21HH331 pKa = 7.24LKK333 pKa = 10.74RR334 pKa = 11.84LWEE337 pKa = 4.07WTRR340 pKa = 11.84EE341 pKa = 3.81ACLRR345 pKa = 11.84MPHH348 pKa = 6.96RR349 pKa = 11.84MMDD352 pKa = 3.44GSVYY356 pKa = 10.2EE357 pKa = 4.08RR358 pKa = 11.84LFRR361 pKa = 11.84CIPSGLFTTQFLDD374 pKa = 3.11SFYY377 pKa = 11.35NMLMILTILSAMGIDD392 pKa = 3.19ISTVVIRR399 pKa = 11.84VQGDD403 pKa = 3.17DD404 pKa = 4.05SLIMLQFFLPANQHH418 pKa = 5.45EE419 pKa = 4.61EE420 pKa = 4.1FKK422 pKa = 11.38ARR424 pKa = 11.84FQALATYY431 pKa = 9.85YY432 pKa = 9.6FDD434 pKa = 5.8HH435 pKa = 7.31IARR438 pKa = 11.84PEE440 pKa = 3.87KK441 pKa = 10.16TDD443 pKa = 3.06ISNSPEE449 pKa = 3.75GVNVLGYY456 pKa = 8.94TNEE459 pKa = 3.78NGYY462 pKa = 9.13PIRR465 pKa = 11.84DD466 pKa = 3.36WRR468 pKa = 11.84KK469 pKa = 9.16LLAQLYY475 pKa = 9.05HH476 pKa = 6.8PRR478 pKa = 11.84SQRR481 pKa = 11.84PTLEE485 pKa = 3.79LLKK488 pKa = 11.02ARR490 pKa = 11.84VCGIQYY496 pKa = 10.88ASMYY500 pKa = 9.76RR501 pKa = 11.84YY502 pKa = 8.52PQVTEE507 pKa = 4.07VCQTLWNRR515 pKa = 11.84LEE517 pKa = 4.09ADD519 pKa = 4.2GIKK522 pKa = 10.52ALNLAAQRR530 pKa = 11.84DD531 pKa = 4.64VVLHH535 pKa = 5.25SHH537 pKa = 7.15ADD539 pKa = 3.45FFIPTDD545 pKa = 3.52HH546 pKa = 7.14FPTLNEE552 pKa = 3.63VTKK555 pKa = 9.72WLRR558 pKa = 11.84VPYY561 pKa = 10.32QRR563 pKa = 11.84TAEE566 pKa = 3.95DD567 pKa = 3.53RR568 pKa = 11.84EE569 pKa = 4.48EE570 pKa = 4.19YY571 pKa = 10.62FPLSHH576 pKa = 6.92FLSYY580 pKa = 10.54FF581 pKa = 3.3
MM1 pKa = 7.53FSLLTWLSTKK11 pKa = 10.18FNALFDD17 pKa = 4.19SVYY20 pKa = 9.57STRR23 pKa = 11.84FEE25 pKa = 4.79HH26 pKa = 6.93NFRR29 pKa = 11.84FLYY32 pKa = 10.05KK33 pKa = 10.18AAPLTTPHH41 pKa = 7.19RR42 pKa = 11.84DD43 pKa = 2.99EE44 pKa = 4.19TKK46 pKa = 8.62YY47 pKa = 11.03AEE49 pKa = 3.93YY50 pKa = 10.0QAYY53 pKa = 9.82VKK55 pKa = 9.91STLDD59 pKa = 3.44HH60 pKa = 6.4VLLGPDD66 pKa = 2.8AHH68 pKa = 6.9HH69 pKa = 6.76IVNGYY74 pKa = 7.96HH75 pKa = 6.58HH76 pKa = 7.84PIATLDD82 pKa = 3.51AVINTFKK89 pKa = 11.0KK90 pKa = 10.79GDD92 pKa = 3.96LPDD95 pKa = 3.67HH96 pKa = 7.09KK97 pKa = 10.43IPKK100 pKa = 8.97DD101 pKa = 3.27LHH103 pKa = 6.49YY104 pKa = 11.16YY105 pKa = 9.9SALLEE110 pKa = 4.3TTKK113 pKa = 10.69RR114 pKa = 11.84FAPPQKK120 pKa = 10.07LRR122 pKa = 11.84PVHH125 pKa = 6.3FADD128 pKa = 4.35LRR130 pKa = 11.84FYY132 pKa = 10.37KK133 pKa = 10.12WNWHH137 pKa = 6.26PNVEE141 pKa = 4.17EE142 pKa = 4.25PFRR145 pKa = 11.84SEE147 pKa = 5.57AEE149 pKa = 3.86LQQAVEE155 pKa = 4.24AAYY158 pKa = 9.87HH159 pKa = 6.64AGLLPDD165 pKa = 3.5GRR167 pKa = 11.84MSFGNLKK174 pKa = 9.49NVVFIRR180 pKa = 11.84VRR182 pKa = 11.84EE183 pKa = 3.89WLHH186 pKa = 4.79QIKK189 pKa = 10.09RR190 pKa = 11.84AQITNPNTLYY200 pKa = 10.29PLINIHH206 pKa = 6.06VKK208 pKa = 9.89PALTTPDD215 pKa = 3.12EE216 pKa = 4.25TKK218 pKa = 10.01IRR220 pKa = 11.84VVYY223 pKa = 10.01GVSKK227 pKa = 10.35LHH229 pKa = 5.83VLPEE233 pKa = 4.17AMFFWPLFRR242 pKa = 11.84HH243 pKa = 5.55YY244 pKa = 10.99LDD246 pKa = 3.84NRR248 pKa = 11.84KK249 pKa = 9.37EE250 pKa = 4.43SPLLWGFEE258 pKa = 4.33TILGGMAVLHH268 pKa = 6.28NMMTIPRR275 pKa = 11.84LYY277 pKa = 8.89FQTFVMVDD285 pKa = 2.48WSGFDD290 pKa = 3.43LRR292 pKa = 11.84SLFEE296 pKa = 4.72PIRR299 pKa = 11.84TDD301 pKa = 3.62VFPAWRR307 pKa = 11.84TYY309 pKa = 10.68FDD311 pKa = 4.42FNNGYY316 pKa = 9.64IPTRR320 pKa = 11.84FYY322 pKa = 10.21RR323 pKa = 11.84TSKK326 pKa = 10.95ADD328 pKa = 3.52PEE330 pKa = 4.21HH331 pKa = 7.24LKK333 pKa = 10.74RR334 pKa = 11.84LWEE337 pKa = 4.07WTRR340 pKa = 11.84EE341 pKa = 3.81ACLRR345 pKa = 11.84MPHH348 pKa = 6.96RR349 pKa = 11.84MMDD352 pKa = 3.44GSVYY356 pKa = 10.2EE357 pKa = 4.08RR358 pKa = 11.84LFRR361 pKa = 11.84CIPSGLFTTQFLDD374 pKa = 3.11SFYY377 pKa = 11.35NMLMILTILSAMGIDD392 pKa = 3.19ISTVVIRR399 pKa = 11.84VQGDD403 pKa = 3.17DD404 pKa = 4.05SLIMLQFFLPANQHH418 pKa = 5.45EE419 pKa = 4.61EE420 pKa = 4.1FKK422 pKa = 11.38ARR424 pKa = 11.84FQALATYY431 pKa = 9.85YY432 pKa = 9.6FDD434 pKa = 5.8HH435 pKa = 7.31IARR438 pKa = 11.84PEE440 pKa = 3.87KK441 pKa = 10.16TDD443 pKa = 3.06ISNSPEE449 pKa = 3.75GVNVLGYY456 pKa = 8.94TNEE459 pKa = 3.78NGYY462 pKa = 9.13PIRR465 pKa = 11.84DD466 pKa = 3.36WRR468 pKa = 11.84KK469 pKa = 9.16LLAQLYY475 pKa = 9.05HH476 pKa = 6.8PRR478 pKa = 11.84SQRR481 pKa = 11.84PTLEE485 pKa = 3.79LLKK488 pKa = 11.02ARR490 pKa = 11.84VCGIQYY496 pKa = 10.88ASMYY500 pKa = 9.76RR501 pKa = 11.84YY502 pKa = 8.52PQVTEE507 pKa = 4.07VCQTLWNRR515 pKa = 11.84LEE517 pKa = 4.09ADD519 pKa = 4.2GIKK522 pKa = 10.52ALNLAAQRR530 pKa = 11.84DD531 pKa = 4.64VVLHH535 pKa = 5.25SHH537 pKa = 7.15ADD539 pKa = 3.45FFIPTDD545 pKa = 3.52HH546 pKa = 7.14FPTLNEE552 pKa = 3.63VTKK555 pKa = 9.72WLRR558 pKa = 11.84VPYY561 pKa = 10.32QRR563 pKa = 11.84TAEE566 pKa = 3.95DD567 pKa = 3.53RR568 pKa = 11.84EE569 pKa = 4.48EE570 pKa = 4.19YY571 pKa = 10.62FPLSHH576 pKa = 6.92FLSYY580 pKa = 10.54FF581 pKa = 3.3
Molecular weight: 68.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1093 |
512 |
581 |
546.5 |
62.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.417 ± 1.496 | 0.549 ± 0.111 |
5.947 ± 0.625 | 4.3 ± 0.826 |
6.404 ± 0.383 | 4.392 ± 0.62 |
3.751 ± 0.577 | 4.849 ± 0.161 |
4.392 ± 0.071 | 9.79 ± 0.977 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.745 ± 0.13 | 5.032 ± 0.719 |
6.404 ± 0.029 | 3.294 ± 0.019 |
5.947 ± 0.748 | 6.038 ± 1.247 |
6.221 ± 0.254 | 5.947 ± 0.213 |
1.555 ± 0.544 | 4.026 ± 0.907 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
