
Longjawed orbweaver circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Circularisvirus
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
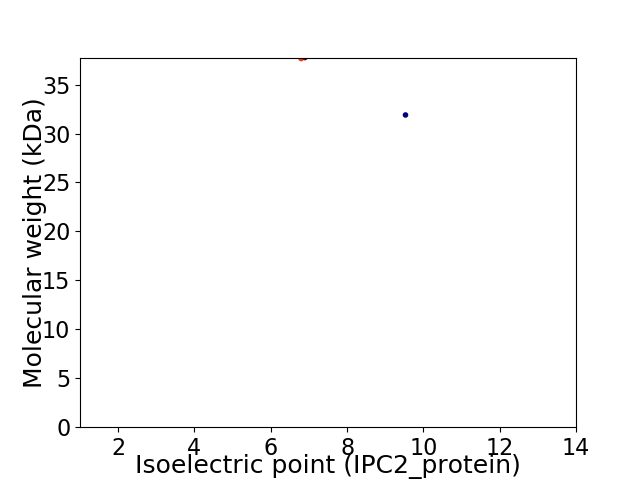
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
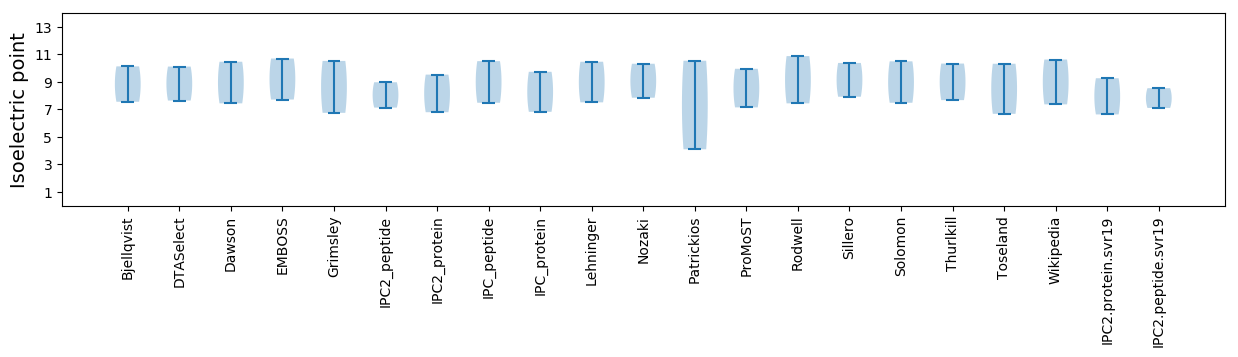
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPA7|A0A346BPA7_9VIRU Putative capsid protein OS=Longjawed orbweaver circular virus 1 OX=2293294 PE=4 SV=1
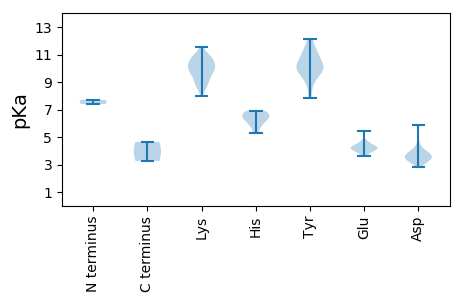
MM1 pKa = 7.4SRR3 pKa = 11.84QKK5 pKa = 10.11GWCYY9 pKa = 10.06TRR11 pKa = 11.84FDD13 pKa = 3.7IEE15 pKa = 4.7NIPVYY20 pKa = 10.45NEE22 pKa = 4.06HH23 pKa = 6.86EE24 pKa = 4.12EE25 pKa = 4.35SYY27 pKa = 10.73HH28 pKa = 6.31IFGRR32 pKa = 11.84EE33 pKa = 3.65RR34 pKa = 11.84CPSTGRR40 pKa = 11.84IHH42 pKa = 5.79YY43 pKa = 10.22QGFIYY48 pKa = 9.91FKK50 pKa = 9.58QRR52 pKa = 11.84KK53 pKa = 8.91RR54 pKa = 11.84LTQLKK59 pKa = 8.72KK60 pKa = 10.55VGYY63 pKa = 7.81TGHH66 pKa = 6.56FEE68 pKa = 4.24AMRR71 pKa = 11.84GSPQQASEE79 pKa = 4.29YY80 pKa = 9.66CKK82 pKa = 10.35KK83 pKa = 10.84DD84 pKa = 2.79SDD86 pKa = 4.03YY87 pKa = 11.71TEE89 pKa = 5.42FGQLPVVSSGGSVFADD105 pKa = 3.46ALAKK109 pKa = 10.59AKK111 pKa = 10.82AGDD114 pKa = 3.58FDD116 pKa = 5.9SIEE119 pKa = 4.18EE120 pKa = 3.97NHH122 pKa = 6.81PGVFLRR128 pKa = 11.84YY129 pKa = 9.49RR130 pKa = 11.84GTLLSCFKK138 pKa = 10.89YY139 pKa = 10.83NVVEE143 pKa = 4.2LLNSCGVWICGPPRR157 pKa = 11.84SGKK160 pKa = 10.17DD161 pKa = 3.15YY162 pKa = 11.03SVRR165 pKa = 11.84KK166 pKa = 9.87LSSLYY171 pKa = 10.13IKK173 pKa = 10.02PLNKK177 pKa = 9.22WWDD180 pKa = 3.51GYY182 pKa = 9.8MNEE185 pKa = 4.42DD186 pKa = 3.31NVLISDD192 pKa = 3.8VDD194 pKa = 3.91VSHH197 pKa = 6.68GNWLGYY203 pKa = 8.57FLKK206 pKa = 10.23IWMDD210 pKa = 3.74RR211 pKa = 11.84YY212 pKa = 11.21AFIGEE217 pKa = 4.2IKK219 pKa = 10.43GGSIKK224 pKa = 10.03IRR226 pKa = 11.84PKK228 pKa = 10.56KK229 pKa = 10.79VFVTSNFLIEE239 pKa = 4.37DD240 pKa = 3.72VFVNEE245 pKa = 4.07QVCAAVKK252 pKa = 10.12ARR254 pKa = 11.84CNIYY258 pKa = 10.7NLFEE262 pKa = 4.26HH263 pKa = 6.35VVTRR267 pKa = 11.84RR268 pKa = 11.84VDD270 pKa = 3.35INPSDD275 pKa = 4.5DD276 pKa = 3.64LFKK279 pKa = 11.14LLEE282 pKa = 4.27EE283 pKa = 4.7NGDD286 pKa = 3.84VVQTSTRR293 pKa = 11.84APLSEE298 pKa = 4.06VQAEE302 pKa = 4.58VVAGPSGHH310 pKa = 6.25VPPKK314 pKa = 9.99KK315 pKa = 8.63KK316 pKa = 10.54CKK318 pKa = 8.51VAEE321 pKa = 4.18VFSDD325 pKa = 4.0SEE327 pKa = 4.26DD328 pKa = 3.81FEE330 pKa = 4.65
MM1 pKa = 7.4SRR3 pKa = 11.84QKK5 pKa = 10.11GWCYY9 pKa = 10.06TRR11 pKa = 11.84FDD13 pKa = 3.7IEE15 pKa = 4.7NIPVYY20 pKa = 10.45NEE22 pKa = 4.06HH23 pKa = 6.86EE24 pKa = 4.12EE25 pKa = 4.35SYY27 pKa = 10.73HH28 pKa = 6.31IFGRR32 pKa = 11.84EE33 pKa = 3.65RR34 pKa = 11.84CPSTGRR40 pKa = 11.84IHH42 pKa = 5.79YY43 pKa = 10.22QGFIYY48 pKa = 9.91FKK50 pKa = 9.58QRR52 pKa = 11.84KK53 pKa = 8.91RR54 pKa = 11.84LTQLKK59 pKa = 8.72KK60 pKa = 10.55VGYY63 pKa = 7.81TGHH66 pKa = 6.56FEE68 pKa = 4.24AMRR71 pKa = 11.84GSPQQASEE79 pKa = 4.29YY80 pKa = 9.66CKK82 pKa = 10.35KK83 pKa = 10.84DD84 pKa = 2.79SDD86 pKa = 4.03YY87 pKa = 11.71TEE89 pKa = 5.42FGQLPVVSSGGSVFADD105 pKa = 3.46ALAKK109 pKa = 10.59AKK111 pKa = 10.82AGDD114 pKa = 3.58FDD116 pKa = 5.9SIEE119 pKa = 4.18EE120 pKa = 3.97NHH122 pKa = 6.81PGVFLRR128 pKa = 11.84YY129 pKa = 9.49RR130 pKa = 11.84GTLLSCFKK138 pKa = 10.89YY139 pKa = 10.83NVVEE143 pKa = 4.2LLNSCGVWICGPPRR157 pKa = 11.84SGKK160 pKa = 10.17DD161 pKa = 3.15YY162 pKa = 11.03SVRR165 pKa = 11.84KK166 pKa = 9.87LSSLYY171 pKa = 10.13IKK173 pKa = 10.02PLNKK177 pKa = 9.22WWDD180 pKa = 3.51GYY182 pKa = 9.8MNEE185 pKa = 4.42DD186 pKa = 3.31NVLISDD192 pKa = 3.8VDD194 pKa = 3.91VSHH197 pKa = 6.68GNWLGYY203 pKa = 8.57FLKK206 pKa = 10.23IWMDD210 pKa = 3.74RR211 pKa = 11.84YY212 pKa = 11.21AFIGEE217 pKa = 4.2IKK219 pKa = 10.43GGSIKK224 pKa = 10.03IRR226 pKa = 11.84PKK228 pKa = 10.56KK229 pKa = 10.79VFVTSNFLIEE239 pKa = 4.37DD240 pKa = 3.72VFVNEE245 pKa = 4.07QVCAAVKK252 pKa = 10.12ARR254 pKa = 11.84CNIYY258 pKa = 10.7NLFEE262 pKa = 4.26HH263 pKa = 6.35VVTRR267 pKa = 11.84RR268 pKa = 11.84VDD270 pKa = 3.35INPSDD275 pKa = 4.5DD276 pKa = 3.64LFKK279 pKa = 11.14LLEE282 pKa = 4.27EE283 pKa = 4.7NGDD286 pKa = 3.84VVQTSTRR293 pKa = 11.84APLSEE298 pKa = 4.06VQAEE302 pKa = 4.58VVAGPSGHH310 pKa = 6.25VPPKK314 pKa = 9.99KK315 pKa = 8.63KK316 pKa = 10.54CKK318 pKa = 8.51VAEE321 pKa = 4.18VFSDD325 pKa = 4.0SEE327 pKa = 4.26DD328 pKa = 3.81FEE330 pKa = 4.65
Molecular weight: 37.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPA7|A0A346BPA7_9VIRU Putative capsid protein OS=Longjawed orbweaver circular virus 1 OX=2293294 PE=4 SV=1
MM1 pKa = 7.71LFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84LGLRR10 pKa = 11.84YY11 pKa = 9.13RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.86RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.32SWPARR22 pKa = 11.84RR23 pKa = 11.84AMYY26 pKa = 9.96RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84SAKK33 pKa = 8.79LRR35 pKa = 11.84KK36 pKa = 8.88SSRR39 pKa = 11.84IRR41 pKa = 11.84RR42 pKa = 11.84ISNDD46 pKa = 3.03YY47 pKa = 9.68TKK49 pKa = 10.06EE50 pKa = 4.13TFSDD54 pKa = 3.12VTMYY58 pKa = 11.27VLLPDD63 pKa = 3.41KK64 pKa = 10.66VKK66 pKa = 10.66PEE68 pKa = 4.3RR69 pKa = 11.84PSSTSFEE76 pKa = 4.38MFTSAIGTDD85 pKa = 3.34SMNSDD90 pKa = 3.28RR91 pKa = 11.84FGFLCTVYY99 pKa = 10.41KK100 pKa = 10.14YY101 pKa = 11.41VKK103 pKa = 9.59FNSFVWHH110 pKa = 5.27VTCDD114 pKa = 3.99PITAVSGAFAVEE126 pKa = 3.92KK127 pKa = 9.63TVVKK131 pKa = 9.63GTVSGLNEE139 pKa = 3.81FFKK142 pKa = 11.55NEE144 pKa = 3.91DD145 pKa = 4.33FYY147 pKa = 12.15VSWDD151 pKa = 3.39LNFAYY156 pKa = 10.07DD157 pKa = 3.44ASKK160 pKa = 11.07AITVKK165 pKa = 10.75DD166 pKa = 3.34LTGIHH171 pKa = 6.44AKK173 pKa = 7.98KK174 pKa = 8.86TRR176 pKa = 11.84IGGHH180 pKa = 5.92KK181 pKa = 9.62PAIFKK186 pKa = 10.12WKK188 pKa = 9.88VPQKK192 pKa = 9.82LRR194 pKa = 11.84EE195 pKa = 3.94YY196 pKa = 9.18VLCSEE201 pKa = 4.55VPDD204 pKa = 3.73ARR206 pKa = 11.84SGNVPIGSFFNNLLEE221 pKa = 4.45RR222 pKa = 11.84KK223 pKa = 9.5NINVPNYY230 pKa = 10.24FIGCGTEE237 pKa = 3.78IMRR240 pKa = 11.84KK241 pKa = 9.4LPNFNNMFDD250 pKa = 3.66DD251 pKa = 3.48VRR253 pKa = 11.84FVLRR257 pKa = 11.84VNCFVNCTFHH267 pKa = 6.91GAVILKK273 pKa = 9.13QQ274 pKa = 3.26
MM1 pKa = 7.71LFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84LGLRR10 pKa = 11.84YY11 pKa = 9.13RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.86RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.32SWPARR22 pKa = 11.84RR23 pKa = 11.84AMYY26 pKa = 9.96RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84SAKK33 pKa = 8.79LRR35 pKa = 11.84KK36 pKa = 8.88SSRR39 pKa = 11.84IRR41 pKa = 11.84RR42 pKa = 11.84ISNDD46 pKa = 3.03YY47 pKa = 9.68TKK49 pKa = 10.06EE50 pKa = 4.13TFSDD54 pKa = 3.12VTMYY58 pKa = 11.27VLLPDD63 pKa = 3.41KK64 pKa = 10.66VKK66 pKa = 10.66PEE68 pKa = 4.3RR69 pKa = 11.84PSSTSFEE76 pKa = 4.38MFTSAIGTDD85 pKa = 3.34SMNSDD90 pKa = 3.28RR91 pKa = 11.84FGFLCTVYY99 pKa = 10.41KK100 pKa = 10.14YY101 pKa = 11.41VKK103 pKa = 9.59FNSFVWHH110 pKa = 5.27VTCDD114 pKa = 3.99PITAVSGAFAVEE126 pKa = 3.92KK127 pKa = 9.63TVVKK131 pKa = 9.63GTVSGLNEE139 pKa = 3.81FFKK142 pKa = 11.55NEE144 pKa = 3.91DD145 pKa = 4.33FYY147 pKa = 12.15VSWDD151 pKa = 3.39LNFAYY156 pKa = 10.07DD157 pKa = 3.44ASKK160 pKa = 11.07AITVKK165 pKa = 10.75DD166 pKa = 3.34LTGIHH171 pKa = 6.44AKK173 pKa = 7.98KK174 pKa = 8.86TRR176 pKa = 11.84IGGHH180 pKa = 5.92KK181 pKa = 9.62PAIFKK186 pKa = 10.12WKK188 pKa = 9.88VPQKK192 pKa = 9.82LRR194 pKa = 11.84EE195 pKa = 3.94YY196 pKa = 9.18VLCSEE201 pKa = 4.55VPDD204 pKa = 3.73ARR206 pKa = 11.84SGNVPIGSFFNNLLEE221 pKa = 4.45RR222 pKa = 11.84KK223 pKa = 9.5NINVPNYY230 pKa = 10.24FIGCGTEE237 pKa = 3.78IMRR240 pKa = 11.84KK241 pKa = 9.4LPNFNNMFDD250 pKa = 3.66DD251 pKa = 3.48VRR253 pKa = 11.84FVLRR257 pKa = 11.84VNCFVNCTFHH267 pKa = 6.91GAVILKK273 pKa = 9.13QQ274 pKa = 3.26
Molecular weight: 31.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
604 |
274 |
330 |
302.0 |
34.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.801 ± 0.19 | 2.483 ± 0.181 |
5.298 ± 0.341 | 5.464 ± 1.119 |
6.788 ± 0.766 | 6.457 ± 0.831 |
1.987 ± 0.325 | 4.967 ± 0.137 |
7.781 ± 0.153 | 6.291 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.821 ± 0.452 | 5.298 ± 0.559 |
4.139 ± 0.077 | 1.987 ± 0.775 |
7.45 ± 1.483 | 7.616 ± 0.195 |
4.305 ± 0.947 | 8.94 ± 0.112 |
1.656 ± 0.121 | 4.47 ± 0.281 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
