
Hubei diptera virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
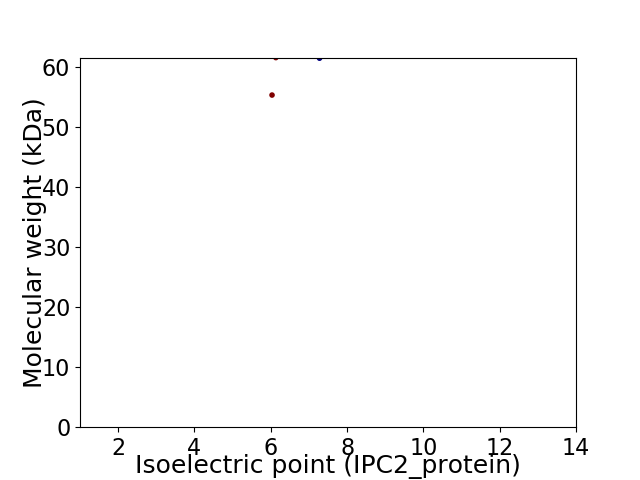
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLM4|A0A1L3KLM4_9VIRU RdRp OS=Hubei diptera virus 18 OX=1922879 PE=4 SV=1
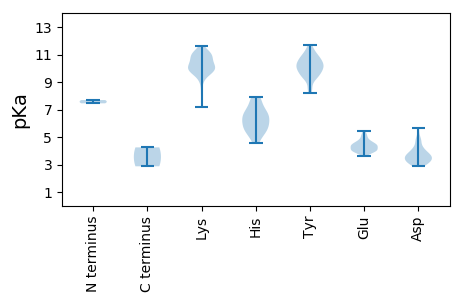
MM1 pKa = 7.45SRR3 pKa = 11.84YY4 pKa = 9.58NFNRR8 pKa = 11.84GAPRR12 pKa = 11.84GRR14 pKa = 11.84GGQWRR19 pKa = 11.84APQNNNYY26 pKa = 9.76DD27 pKa = 2.98AGYY30 pKa = 9.8YY31 pKa = 9.6QNQNQNNRR39 pKa = 11.84SNNNYY44 pKa = 9.52PEE46 pKa = 4.47NNSYY50 pKa = 9.82MYY52 pKa = 8.6EE53 pKa = 4.07APQTSGRR60 pKa = 11.84GRR62 pKa = 11.84GRR64 pKa = 11.84GGYY67 pKa = 9.33RR68 pKa = 11.84RR69 pKa = 11.84GGISNDD75 pKa = 2.9PKK77 pKa = 10.88VNVRR81 pKa = 11.84NPLDD85 pKa = 3.82AKK87 pKa = 11.21DD88 pKa = 4.05PVAQGKK94 pKa = 7.16PASMPTGQHH103 pKa = 5.48LSKK106 pKa = 10.12TNSEE110 pKa = 4.5LKK112 pKa = 10.42NDD114 pKa = 4.26PLYY117 pKa = 11.34LEE119 pKa = 5.42AFFPSPSEE127 pKa = 3.8EE128 pKa = 3.93LVPLPEE134 pKa = 4.38EE135 pKa = 4.45LNHH138 pKa = 5.71FTGFDD143 pKa = 3.57GLSSIINEE151 pKa = 4.47SYY153 pKa = 11.68DD154 pKa = 3.58NFCSHH159 pKa = 6.66SLNFKK164 pKa = 10.36RR165 pKa = 11.84SVSLSSYY172 pKa = 10.34AYY174 pKa = 9.8YY175 pKa = 10.27ISVFCWARR183 pKa = 11.84VLHH186 pKa = 6.5LKK188 pKa = 10.27RR189 pKa = 11.84LNKK192 pKa = 9.77YY193 pKa = 10.05RR194 pKa = 11.84LTTNEE199 pKa = 3.76IEE201 pKa = 4.37FVDD204 pKa = 4.28MIYY207 pKa = 10.9NQGNYY212 pKa = 10.15LLPKK216 pKa = 10.31SVTLYY221 pKa = 11.1LSGFGNFTIPSSVEE235 pKa = 3.94SKK237 pKa = 11.04FNTKK241 pKa = 9.99PYY243 pKa = 10.29SYY245 pKa = 10.99DD246 pKa = 3.23DD247 pKa = 3.22AGYY250 pKa = 9.37FVNFAEE256 pKa = 4.32EE257 pKa = 5.2FYY259 pKa = 10.67TVTSYY264 pKa = 11.36PCISIFAEE272 pKa = 4.24RR273 pKa = 11.84IMRR276 pKa = 11.84DD277 pKa = 3.04MAFTEE282 pKa = 4.01NNRR285 pKa = 11.84VGEE288 pKa = 4.11AWSPNDD294 pKa = 5.67IEE296 pKa = 5.04QEE298 pKa = 3.98WNTRR302 pKa = 11.84CLGYY306 pKa = 10.65APSVEE311 pKa = 4.38LPSLQLQILNMAGISSEE328 pKa = 4.19NFPSDD333 pKa = 4.83CEE335 pKa = 3.97GLLINIRR342 pKa = 11.84LLNSIQKK349 pKa = 9.58YY350 pKa = 9.8LLEE353 pKa = 4.67IPSFEE358 pKa = 4.69SGPIPANLTGSLGQFVVEE376 pKa = 4.04EE377 pKa = 4.26VVEE380 pKa = 4.09NAVALDD386 pKa = 4.56AIDD389 pKa = 4.77NIGSASFITKK399 pKa = 10.21SPLSCPGSLSYY410 pKa = 11.08LGGSFLYY417 pKa = 10.14RR418 pKa = 11.84VAKK421 pKa = 10.07KK422 pKa = 10.21ALPLNKK428 pKa = 9.41IKK430 pKa = 10.7FFFPYY435 pKa = 9.48TINDD439 pKa = 3.34INQLDD444 pKa = 4.12INRR447 pKa = 11.84LDD449 pKa = 3.86TLNHH453 pKa = 5.58GWSPLLEE460 pKa = 4.66NIYY463 pKa = 10.39HH464 pKa = 5.8YY465 pKa = 11.54SNVQFKK471 pKa = 10.57PILRR475 pKa = 11.84LKK477 pKa = 9.61KK478 pKa = 9.99FCSIDD483 pKa = 3.54VKK485 pKa = 9.95PTSVV489 pKa = 2.87
MM1 pKa = 7.45SRR3 pKa = 11.84YY4 pKa = 9.58NFNRR8 pKa = 11.84GAPRR12 pKa = 11.84GRR14 pKa = 11.84GGQWRR19 pKa = 11.84APQNNNYY26 pKa = 9.76DD27 pKa = 2.98AGYY30 pKa = 9.8YY31 pKa = 9.6QNQNQNNRR39 pKa = 11.84SNNNYY44 pKa = 9.52PEE46 pKa = 4.47NNSYY50 pKa = 9.82MYY52 pKa = 8.6EE53 pKa = 4.07APQTSGRR60 pKa = 11.84GRR62 pKa = 11.84GRR64 pKa = 11.84GGYY67 pKa = 9.33RR68 pKa = 11.84RR69 pKa = 11.84GGISNDD75 pKa = 2.9PKK77 pKa = 10.88VNVRR81 pKa = 11.84NPLDD85 pKa = 3.82AKK87 pKa = 11.21DD88 pKa = 4.05PVAQGKK94 pKa = 7.16PASMPTGQHH103 pKa = 5.48LSKK106 pKa = 10.12TNSEE110 pKa = 4.5LKK112 pKa = 10.42NDD114 pKa = 4.26PLYY117 pKa = 11.34LEE119 pKa = 5.42AFFPSPSEE127 pKa = 3.8EE128 pKa = 3.93LVPLPEE134 pKa = 4.38EE135 pKa = 4.45LNHH138 pKa = 5.71FTGFDD143 pKa = 3.57GLSSIINEE151 pKa = 4.47SYY153 pKa = 11.68DD154 pKa = 3.58NFCSHH159 pKa = 6.66SLNFKK164 pKa = 10.36RR165 pKa = 11.84SVSLSSYY172 pKa = 10.34AYY174 pKa = 9.8YY175 pKa = 10.27ISVFCWARR183 pKa = 11.84VLHH186 pKa = 6.5LKK188 pKa = 10.27RR189 pKa = 11.84LNKK192 pKa = 9.77YY193 pKa = 10.05RR194 pKa = 11.84LTTNEE199 pKa = 3.76IEE201 pKa = 4.37FVDD204 pKa = 4.28MIYY207 pKa = 10.9NQGNYY212 pKa = 10.15LLPKK216 pKa = 10.31SVTLYY221 pKa = 11.1LSGFGNFTIPSSVEE235 pKa = 3.94SKK237 pKa = 11.04FNTKK241 pKa = 9.99PYY243 pKa = 10.29SYY245 pKa = 10.99DD246 pKa = 3.23DD247 pKa = 3.22AGYY250 pKa = 9.37FVNFAEE256 pKa = 4.32EE257 pKa = 5.2FYY259 pKa = 10.67TVTSYY264 pKa = 11.36PCISIFAEE272 pKa = 4.24RR273 pKa = 11.84IMRR276 pKa = 11.84DD277 pKa = 3.04MAFTEE282 pKa = 4.01NNRR285 pKa = 11.84VGEE288 pKa = 4.11AWSPNDD294 pKa = 5.67IEE296 pKa = 5.04QEE298 pKa = 3.98WNTRR302 pKa = 11.84CLGYY306 pKa = 10.65APSVEE311 pKa = 4.38LPSLQLQILNMAGISSEE328 pKa = 4.19NFPSDD333 pKa = 4.83CEE335 pKa = 3.97GLLINIRR342 pKa = 11.84LLNSIQKK349 pKa = 9.58YY350 pKa = 9.8LLEE353 pKa = 4.67IPSFEE358 pKa = 4.69SGPIPANLTGSLGQFVVEE376 pKa = 4.04EE377 pKa = 4.26VVEE380 pKa = 4.09NAVALDD386 pKa = 4.56AIDD389 pKa = 4.77NIGSASFITKK399 pKa = 10.21SPLSCPGSLSYY410 pKa = 11.08LGGSFLYY417 pKa = 10.14RR418 pKa = 11.84VAKK421 pKa = 10.07KK422 pKa = 10.21ALPLNKK428 pKa = 9.41IKK430 pKa = 10.7FFFPYY435 pKa = 9.48TINDD439 pKa = 3.34INQLDD444 pKa = 4.12INRR447 pKa = 11.84LDD449 pKa = 3.86TLNHH453 pKa = 5.58GWSPLLEE460 pKa = 4.66NIYY463 pKa = 10.39HH464 pKa = 5.8YY465 pKa = 11.54SNVQFKK471 pKa = 10.57PILRR475 pKa = 11.84LKK477 pKa = 9.61KK478 pKa = 9.99FCSIDD483 pKa = 3.54VKK485 pKa = 9.95PTSVV489 pKa = 2.87
Molecular weight: 55.27 kDa
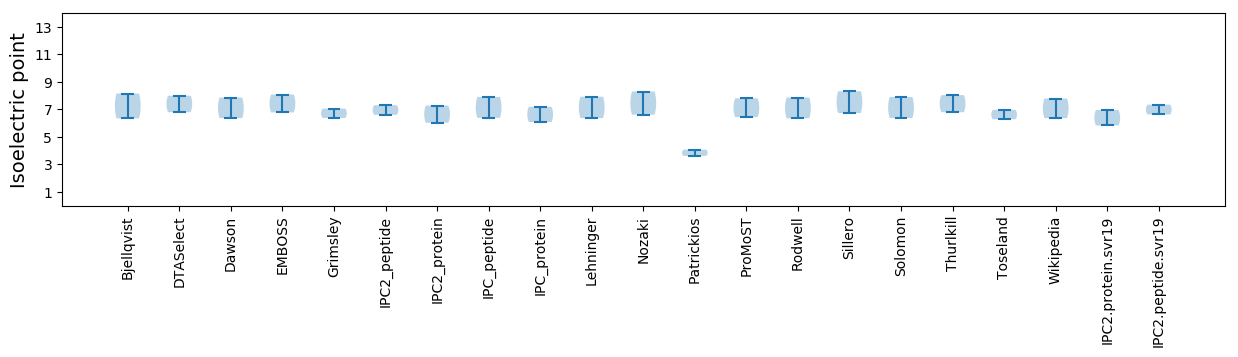
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLM4|A0A1L3KLM4_9VIRU RdRp OS=Hubei diptera virus 18 OX=1922879 PE=4 SV=1
MM1 pKa = 7.69RR2 pKa = 11.84NVTKK6 pKa = 10.5AIRR9 pKa = 11.84EE10 pKa = 3.95LAPAQGYY17 pKa = 8.19VMNMTPARR25 pKa = 11.84EE26 pKa = 4.02DD27 pKa = 3.03GKK29 pKa = 11.15ARR31 pKa = 11.84LACEE35 pKa = 4.08LFFGKK40 pKa = 10.37DD41 pKa = 3.01KK42 pKa = 11.05VDD44 pKa = 3.2KK45 pKa = 9.91VTNTLHH51 pKa = 7.69RR52 pKa = 11.84SRR54 pKa = 11.84LNYY57 pKa = 10.34DD58 pKa = 3.39SLIEE62 pKa = 5.22DD63 pKa = 3.67FLEE66 pKa = 3.8YY67 pKa = 10.84DD68 pKa = 3.37RR69 pKa = 11.84DD70 pKa = 3.59HH71 pKa = 6.52VRR73 pKa = 11.84RR74 pKa = 11.84FEE76 pKa = 5.04DD77 pKa = 4.18PDD79 pKa = 3.5TKK81 pKa = 10.9LIYY84 pKa = 9.72NTVLQSIIDD93 pKa = 3.84DD94 pKa = 4.79LGDD97 pKa = 3.55QRR99 pKa = 11.84IKK101 pKa = 10.82PLTFDD106 pKa = 3.27QVRR109 pKa = 11.84NLSDD113 pKa = 3.65FPGAKK118 pKa = 9.96SPGLPYY124 pKa = 10.51KK125 pKa = 9.97NQGYY129 pKa = 9.22RR130 pKa = 11.84SKK132 pKa = 11.63AEE134 pKa = 3.96VYY136 pKa = 9.85DD137 pKa = 3.75CKK139 pKa = 11.19EE140 pKa = 3.92NLEE143 pKa = 4.12HH144 pKa = 7.59LEE146 pKa = 4.52NIWDD150 pKa = 4.75LIGNGKK156 pKa = 8.95NVYY159 pKa = 10.35LPDD162 pKa = 3.08VCLFARR168 pKa = 11.84SQIAKK173 pKa = 9.71YY174 pKa = 9.24PKK176 pKa = 9.64EE177 pKa = 4.41KK178 pKa = 10.08IRR180 pKa = 11.84ATWGYY185 pKa = 10.4SFDD188 pKa = 3.69VYY190 pKa = 10.53MEE192 pKa = 4.19EE193 pKa = 4.97ARR195 pKa = 11.84FFYY198 pKa = 10.03PIQEE202 pKa = 4.4FIKK205 pKa = 9.75SHH207 pKa = 4.53KK208 pKa = 9.57HH209 pKa = 5.21KK210 pKa = 11.07LPIAYY215 pKa = 9.27GLEE218 pKa = 4.15MAHH221 pKa = 6.94GGMNSINDD229 pKa = 3.38MLLRR233 pKa = 11.84NRR235 pKa = 11.84GCKK238 pKa = 9.87YY239 pKa = 10.8VISDD243 pKa = 3.14WSKK246 pKa = 9.92FDD248 pKa = 3.25KK249 pKa = 10.72TIPPWLIRR257 pKa = 11.84DD258 pKa = 3.75AFHH261 pKa = 7.92ILEE264 pKa = 4.31KK265 pKa = 10.92LIDD268 pKa = 4.12FEE270 pKa = 4.7SLPRR274 pKa = 11.84PAYY277 pKa = 8.42QRR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84FKK283 pKa = 10.93KK284 pKa = 8.17IVDD287 pKa = 3.94YY288 pKa = 10.66FVEE291 pKa = 4.67TPIRR295 pKa = 11.84TCKK298 pKa = 10.63GEE300 pKa = 3.98RR301 pKa = 11.84FLVTGGVPSGSCFTNIIDD319 pKa = 5.23SIINCIVTRR328 pKa = 11.84FLVYY332 pKa = 9.27QTTNQFPIGEE342 pKa = 4.44IFLGDD347 pKa = 3.59DD348 pKa = 3.73GVFVISGFACLEE360 pKa = 4.8DD361 pKa = 3.6IASLAIKK368 pKa = 9.83YY369 pKa = 10.19FGMILNVDD377 pKa = 3.4KK378 pKa = 11.19SYY380 pKa = 10.2VTTNPQNVHH389 pKa = 5.18FLGYY393 pKa = 10.06FNYY396 pKa = 10.91SGMPFKK402 pKa = 11.23NQDD405 pKa = 3.14FLIASFIFPEE415 pKa = 4.29HH416 pKa = 6.37KK417 pKa = 9.12RR418 pKa = 11.84TRR420 pKa = 11.84LIDD423 pKa = 3.39ACAAALGQMYY433 pKa = 10.5SGFDD437 pKa = 3.16PGYY440 pKa = 10.04ARR442 pKa = 11.84TWLKK446 pKa = 10.21IIHH449 pKa = 6.09YY450 pKa = 10.03LADD453 pKa = 3.98AEE455 pKa = 4.33NANPFNFHH463 pKa = 6.93EE464 pKa = 4.73IVLHH468 pKa = 5.73LRR470 pKa = 11.84NNEE473 pKa = 3.66FRR475 pKa = 11.84HH476 pKa = 6.22KK477 pKa = 10.72YY478 pKa = 8.54LAQVGLTSDD487 pKa = 2.95NMTIPEE493 pKa = 4.48LHH495 pKa = 6.74SSMILEE501 pKa = 4.47VLPKK505 pKa = 9.8SYY507 pKa = 10.73CAISLPNRR515 pKa = 11.84TYY517 pKa = 10.81DD518 pKa = 3.51YY519 pKa = 10.04KK520 pKa = 11.07SLYY523 pKa = 9.48IRR525 pKa = 11.84SLSSFPP531 pKa = 4.25
MM1 pKa = 7.69RR2 pKa = 11.84NVTKK6 pKa = 10.5AIRR9 pKa = 11.84EE10 pKa = 3.95LAPAQGYY17 pKa = 8.19VMNMTPARR25 pKa = 11.84EE26 pKa = 4.02DD27 pKa = 3.03GKK29 pKa = 11.15ARR31 pKa = 11.84LACEE35 pKa = 4.08LFFGKK40 pKa = 10.37DD41 pKa = 3.01KK42 pKa = 11.05VDD44 pKa = 3.2KK45 pKa = 9.91VTNTLHH51 pKa = 7.69RR52 pKa = 11.84SRR54 pKa = 11.84LNYY57 pKa = 10.34DD58 pKa = 3.39SLIEE62 pKa = 5.22DD63 pKa = 3.67FLEE66 pKa = 3.8YY67 pKa = 10.84DD68 pKa = 3.37RR69 pKa = 11.84DD70 pKa = 3.59HH71 pKa = 6.52VRR73 pKa = 11.84RR74 pKa = 11.84FEE76 pKa = 5.04DD77 pKa = 4.18PDD79 pKa = 3.5TKK81 pKa = 10.9LIYY84 pKa = 9.72NTVLQSIIDD93 pKa = 3.84DD94 pKa = 4.79LGDD97 pKa = 3.55QRR99 pKa = 11.84IKK101 pKa = 10.82PLTFDD106 pKa = 3.27QVRR109 pKa = 11.84NLSDD113 pKa = 3.65FPGAKK118 pKa = 9.96SPGLPYY124 pKa = 10.51KK125 pKa = 9.97NQGYY129 pKa = 9.22RR130 pKa = 11.84SKK132 pKa = 11.63AEE134 pKa = 3.96VYY136 pKa = 9.85DD137 pKa = 3.75CKK139 pKa = 11.19EE140 pKa = 3.92NLEE143 pKa = 4.12HH144 pKa = 7.59LEE146 pKa = 4.52NIWDD150 pKa = 4.75LIGNGKK156 pKa = 8.95NVYY159 pKa = 10.35LPDD162 pKa = 3.08VCLFARR168 pKa = 11.84SQIAKK173 pKa = 9.71YY174 pKa = 9.24PKK176 pKa = 9.64EE177 pKa = 4.41KK178 pKa = 10.08IRR180 pKa = 11.84ATWGYY185 pKa = 10.4SFDD188 pKa = 3.69VYY190 pKa = 10.53MEE192 pKa = 4.19EE193 pKa = 4.97ARR195 pKa = 11.84FFYY198 pKa = 10.03PIQEE202 pKa = 4.4FIKK205 pKa = 9.75SHH207 pKa = 4.53KK208 pKa = 9.57HH209 pKa = 5.21KK210 pKa = 11.07LPIAYY215 pKa = 9.27GLEE218 pKa = 4.15MAHH221 pKa = 6.94GGMNSINDD229 pKa = 3.38MLLRR233 pKa = 11.84NRR235 pKa = 11.84GCKK238 pKa = 9.87YY239 pKa = 10.8VISDD243 pKa = 3.14WSKK246 pKa = 9.92FDD248 pKa = 3.25KK249 pKa = 10.72TIPPWLIRR257 pKa = 11.84DD258 pKa = 3.75AFHH261 pKa = 7.92ILEE264 pKa = 4.31KK265 pKa = 10.92LIDD268 pKa = 4.12FEE270 pKa = 4.7SLPRR274 pKa = 11.84PAYY277 pKa = 8.42QRR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84FKK283 pKa = 10.93KK284 pKa = 8.17IVDD287 pKa = 3.94YY288 pKa = 10.66FVEE291 pKa = 4.67TPIRR295 pKa = 11.84TCKK298 pKa = 10.63GEE300 pKa = 3.98RR301 pKa = 11.84FLVTGGVPSGSCFTNIIDD319 pKa = 5.23SIINCIVTRR328 pKa = 11.84FLVYY332 pKa = 9.27QTTNQFPIGEE342 pKa = 4.44IFLGDD347 pKa = 3.59DD348 pKa = 3.73GVFVISGFACLEE360 pKa = 4.8DD361 pKa = 3.6IASLAIKK368 pKa = 9.83YY369 pKa = 10.19FGMILNVDD377 pKa = 3.4KK378 pKa = 11.19SYY380 pKa = 10.2VTTNPQNVHH389 pKa = 5.18FLGYY393 pKa = 10.06FNYY396 pKa = 10.91SGMPFKK402 pKa = 11.23NQDD405 pKa = 3.14FLIASFIFPEE415 pKa = 4.29HH416 pKa = 6.37KK417 pKa = 9.12RR418 pKa = 11.84TRR420 pKa = 11.84LIDD423 pKa = 3.39ACAAALGQMYY433 pKa = 10.5SGFDD437 pKa = 3.16PGYY440 pKa = 10.04ARR442 pKa = 11.84TWLKK446 pKa = 10.21IIHH449 pKa = 6.09YY450 pKa = 10.03LADD453 pKa = 3.98AEE455 pKa = 4.33NANPFNFHH463 pKa = 6.93EE464 pKa = 4.73IVLHH468 pKa = 5.73LRR470 pKa = 11.84NNEE473 pKa = 3.66FRR475 pKa = 11.84HH476 pKa = 6.22KK477 pKa = 10.72YY478 pKa = 8.54LAQVGLTSDD487 pKa = 2.95NMTIPEE493 pKa = 4.48LHH495 pKa = 6.74SSMILEE501 pKa = 4.47VLPKK505 pKa = 9.8SYY507 pKa = 10.73CAISLPNRR515 pKa = 11.84TYY517 pKa = 10.81DD518 pKa = 3.51YY519 pKa = 10.04KK520 pKa = 11.07SLYY523 pKa = 9.48IRR525 pKa = 11.84SLSSFPP531 pKa = 4.25
Molecular weight: 61.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1020 |
489 |
531 |
510.0 |
58.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.294 ± 0.217 | 1.667 ± 0.132 |
5.294 ± 0.79 | 5.392 ± 0.187 |
6.078 ± 0.312 | 5.98 ± 0.316 |
1.961 ± 0.412 | 6.863 ± 0.638 |
5.294 ± 0.561 | 9.216 ± 0.107 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.863 ± 0.242 | 7.549 ± 1.271 |
5.784 ± 0.426 | 2.941 ± 0.186 |
5.49 ± 0.327 | 7.843 ± 1.221 |
4.02 ± 0.19 | 4.804 ± 0.056 |
0.98 ± 0.024 | 5.686 ± 0.137 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
