
Roseomonas sp. Z24
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Roseomonas
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
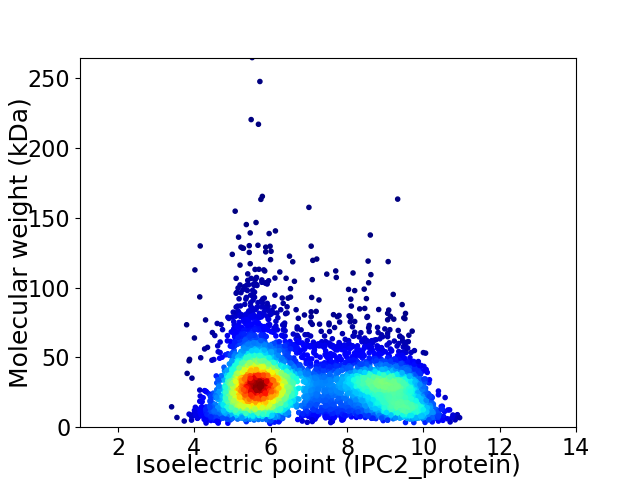
Virtual 2D-PAGE plot for 5527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9JJI7|A0A3A9JJI7_9PROT Mannitol dehydrogenase family protein (Fragment) OS=Roseomonas sp. Z24 OX=2382222 GN=D6Z83_07825 PE=4 SV=1
MM1 pKa = 7.81PNLASPDD8 pKa = 3.71YY9 pKa = 9.83PEE11 pKa = 4.07NAIVLHH17 pKa = 6.64AGDD20 pKa = 4.96NIQEE24 pKa = 4.12AVQAGGEE31 pKa = 4.3GAVFWLEE38 pKa = 3.61AGVYY42 pKa = 9.81RR43 pKa = 11.84MQQITPLEE51 pKa = 4.38GQSFLGASGATLNGARR67 pKa = 11.84LLTEE71 pKa = 4.11FTRR74 pKa = 11.84IDD76 pKa = 3.55GHH78 pKa = 6.08WAAEE82 pKa = 4.14GQTQQGEE89 pKa = 4.13RR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 4.14TDD94 pKa = 2.94EE95 pKa = 4.36SAEE98 pKa = 4.1GAQRR102 pKa = 11.84PGYY105 pKa = 9.25PDD107 pKa = 3.05AVYY110 pKa = 11.34VNDD113 pKa = 3.98QPLTPVDD120 pKa = 5.23SIDD123 pKa = 3.42QLKK126 pKa = 10.07PGTYY130 pKa = 10.16FFDD133 pKa = 3.69YY134 pKa = 10.91DD135 pKa = 3.36ADD137 pKa = 4.8RR138 pKa = 11.84IFFGDD143 pKa = 3.95DD144 pKa = 3.04PTGEE148 pKa = 4.22KK149 pKa = 10.49VEE151 pKa = 4.44AAVSPYY157 pKa = 10.8AIASTANDD165 pKa = 3.41VTVSGLTVEE174 pKa = 5.61KK175 pKa = 10.64YY176 pKa = 7.66ATPTQEE182 pKa = 4.24AAISGNGEE190 pKa = 3.61NWVVSNNEE198 pKa = 3.06VRR200 pKa = 11.84LNYY203 pKa = 10.38GVGINVGDD211 pKa = 3.9GGKK214 pKa = 10.06IMDD217 pKa = 4.9NFVHH221 pKa = 7.65DD222 pKa = 4.37NGQMGLNAGGKK233 pKa = 9.55DD234 pKa = 3.36VLVSGNEE241 pKa = 3.66IASNGFWSGIDD252 pKa = 3.63VLWEE256 pKa = 3.95GGGAKK261 pKa = 8.82FTEE264 pKa = 4.73TEE266 pKa = 3.81GLMVRR271 pKa = 11.84NNYY274 pKa = 9.89SHH276 pKa = 7.68DD277 pKa = 3.77NNGYY281 pKa = 9.17GLWTDD286 pKa = 3.01IDD288 pKa = 4.21NVNTTYY294 pKa = 10.52EE295 pKa = 4.29GNRR298 pKa = 11.84IEE300 pKa = 4.19YY301 pKa = 10.37NSAGGINHH309 pKa = 7.44EE310 pKa = 4.09ISYY313 pKa = 10.26AASIHH318 pKa = 6.74DD319 pKa = 3.71NSFVGNGGNGLTWLWGSAIQVQNSQGVEE347 pKa = 3.69IYY349 pKa = 11.26NNFVDD354 pKa = 5.3ASSGGNGIGLIQQEE368 pKa = 4.33RR369 pKa = 11.84GEE371 pKa = 4.52GAFGDD376 pKa = 4.19YY377 pKa = 9.2VTVNNSVHH385 pKa = 7.34DD386 pKa = 3.84NTLILSSEE394 pKa = 4.27SGVGAVADD402 pKa = 3.87YY403 pKa = 11.4DD404 pKa = 4.28EE405 pKa = 5.04EE406 pKa = 5.35LMLAGGNTFNNNEE419 pKa = 4.13YY420 pKa = 10.17QVQADD425 pKa = 3.68EE426 pKa = 4.6DD427 pKa = 3.75QFAFGDD433 pKa = 4.4YY434 pKa = 9.05YY435 pKa = 11.24TFGDD439 pKa = 4.2FVNVSGQDD447 pKa = 2.98AGSALLIGG455 pKa = 4.33
MM1 pKa = 7.81PNLASPDD8 pKa = 3.71YY9 pKa = 9.83PEE11 pKa = 4.07NAIVLHH17 pKa = 6.64AGDD20 pKa = 4.96NIQEE24 pKa = 4.12AVQAGGEE31 pKa = 4.3GAVFWLEE38 pKa = 3.61AGVYY42 pKa = 9.81RR43 pKa = 11.84MQQITPLEE51 pKa = 4.38GQSFLGASGATLNGARR67 pKa = 11.84LLTEE71 pKa = 4.11FTRR74 pKa = 11.84IDD76 pKa = 3.55GHH78 pKa = 6.08WAAEE82 pKa = 4.14GQTQQGEE89 pKa = 4.13RR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 4.14TDD94 pKa = 2.94EE95 pKa = 4.36SAEE98 pKa = 4.1GAQRR102 pKa = 11.84PGYY105 pKa = 9.25PDD107 pKa = 3.05AVYY110 pKa = 11.34VNDD113 pKa = 3.98QPLTPVDD120 pKa = 5.23SIDD123 pKa = 3.42QLKK126 pKa = 10.07PGTYY130 pKa = 10.16FFDD133 pKa = 3.69YY134 pKa = 10.91DD135 pKa = 3.36ADD137 pKa = 4.8RR138 pKa = 11.84IFFGDD143 pKa = 3.95DD144 pKa = 3.04PTGEE148 pKa = 4.22KK149 pKa = 10.49VEE151 pKa = 4.44AAVSPYY157 pKa = 10.8AIASTANDD165 pKa = 3.41VTVSGLTVEE174 pKa = 5.61KK175 pKa = 10.64YY176 pKa = 7.66ATPTQEE182 pKa = 4.24AAISGNGEE190 pKa = 3.61NWVVSNNEE198 pKa = 3.06VRR200 pKa = 11.84LNYY203 pKa = 10.38GVGINVGDD211 pKa = 3.9GGKK214 pKa = 10.06IMDD217 pKa = 4.9NFVHH221 pKa = 7.65DD222 pKa = 4.37NGQMGLNAGGKK233 pKa = 9.55DD234 pKa = 3.36VLVSGNEE241 pKa = 3.66IASNGFWSGIDD252 pKa = 3.63VLWEE256 pKa = 3.95GGGAKK261 pKa = 8.82FTEE264 pKa = 4.73TEE266 pKa = 3.81GLMVRR271 pKa = 11.84NNYY274 pKa = 9.89SHH276 pKa = 7.68DD277 pKa = 3.77NNGYY281 pKa = 9.17GLWTDD286 pKa = 3.01IDD288 pKa = 4.21NVNTTYY294 pKa = 10.52EE295 pKa = 4.29GNRR298 pKa = 11.84IEE300 pKa = 4.19YY301 pKa = 10.37NSAGGINHH309 pKa = 7.44EE310 pKa = 4.09ISYY313 pKa = 10.26AASIHH318 pKa = 6.74DD319 pKa = 3.71NSFVGNGGNGLTWLWGSAIQVQNSQGVEE347 pKa = 3.69IYY349 pKa = 11.26NNFVDD354 pKa = 5.3ASSGGNGIGLIQQEE368 pKa = 4.33RR369 pKa = 11.84GEE371 pKa = 4.52GAFGDD376 pKa = 4.19YY377 pKa = 9.2VTVNNSVHH385 pKa = 7.34DD386 pKa = 3.84NTLILSSEE394 pKa = 4.27SGVGAVADD402 pKa = 3.87YY403 pKa = 11.4DD404 pKa = 4.28EE405 pKa = 5.04EE406 pKa = 5.35LMLAGGNTFNNNEE419 pKa = 4.13YY420 pKa = 10.17QVQADD425 pKa = 3.68EE426 pKa = 4.6DD427 pKa = 3.75QFAFGDD433 pKa = 4.4YY434 pKa = 9.05YY435 pKa = 11.24TFGDD439 pKa = 4.2FVNVSGQDD447 pKa = 2.98AGSALLIGG455 pKa = 4.33
Molecular weight: 48.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9JI02|A0A3A9JI02_9PROT ABC transporter substrate-binding protein OS=Roseomonas sp. Z24 OX=2382222 GN=D6Z83_09635 PE=4 SV=1
GG1 pKa = 6.95SRR3 pKa = 11.84PRR5 pKa = 11.84PGRR8 pKa = 11.84RR9 pKa = 11.84CPAPPRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84SPASIGQKK31 pKa = 9.57RR32 pKa = 11.84RR33 pKa = 11.84PATGGPAKK41 pKa = 10.3ARR43 pKa = 11.84RR44 pKa = 11.84LPAEE48 pKa = 3.87RR49 pKa = 11.84RR50 pKa = 11.84ANAARR55 pKa = 11.84AGRR58 pKa = 11.84GAAWW62 pKa = 3.28
GG1 pKa = 6.95SRR3 pKa = 11.84PRR5 pKa = 11.84PGRR8 pKa = 11.84RR9 pKa = 11.84CPAPPRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84SPASIGQKK31 pKa = 9.57RR32 pKa = 11.84RR33 pKa = 11.84PATGGPAKK41 pKa = 10.3ARR43 pKa = 11.84RR44 pKa = 11.84LPAEE48 pKa = 3.87RR49 pKa = 11.84RR50 pKa = 11.84ANAARR55 pKa = 11.84AGRR58 pKa = 11.84GAAWW62 pKa = 3.28
Molecular weight: 6.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1684659 |
25 |
2457 |
304.8 |
32.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.186 ± 0.053 | 0.834 ± 0.01 |
4.93 ± 0.019 | 5.616 ± 0.026 |
3.313 ± 0.02 | 9.075 ± 0.025 |
2.061 ± 0.017 | 4.106 ± 0.028 |
2.197 ± 0.024 | 11.206 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.505 ± 0.015 | 2.131 ± 0.019 |
6.18 ± 0.03 | 3.31 ± 0.019 |
8.066 ± 0.033 | 4.785 ± 0.022 |
4.907 ± 0.019 | 7.279 ± 0.026 |
1.477 ± 0.014 | 1.836 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
