
Fusarium redolens polymycovirus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus
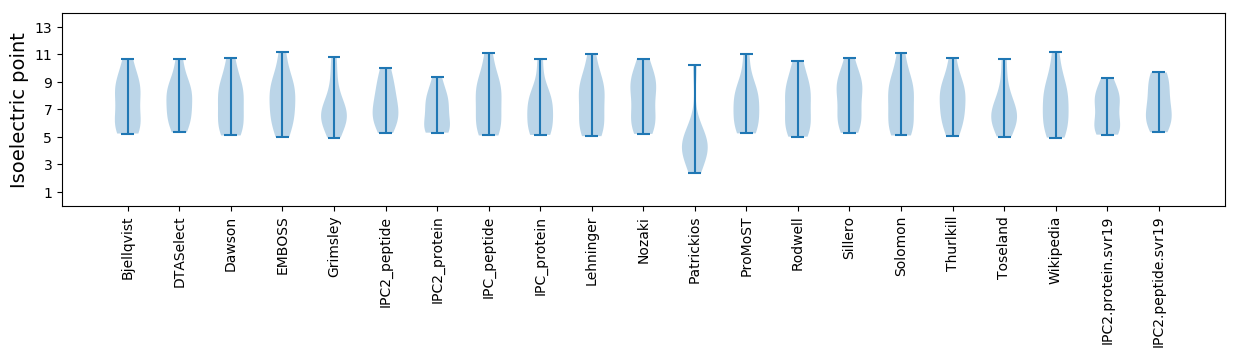
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
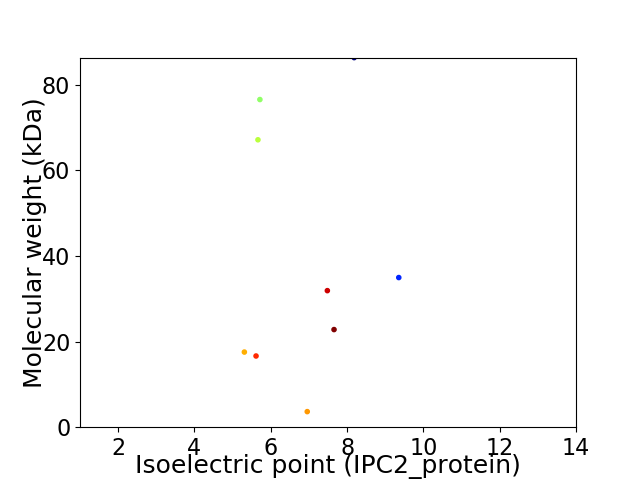
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A513ZVE3|A0A513ZVE3_9VIRU Uncharacterized protein OS=Fusarium redolens polymycovirus 1 OX=2546034 PE=4 SV=1
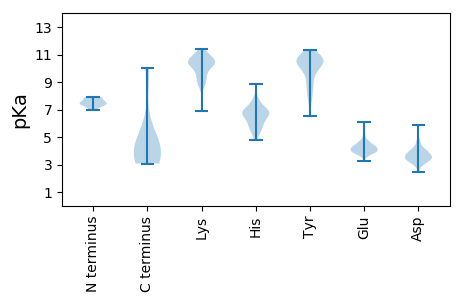
MM1 pKa = 7.34SGKK4 pKa = 10.33SFVIQTEE11 pKa = 4.31CEE13 pKa = 4.19GVGRR17 pKa = 11.84RR18 pKa = 11.84GLVSASVMEE27 pKa = 4.68FYY29 pKa = 10.84FIIGVVEE36 pKa = 4.41DD37 pKa = 3.71VCQEE41 pKa = 3.87VATRR45 pKa = 11.84LACLEE50 pKa = 4.31AEE52 pKa = 4.59FKK54 pKa = 11.01EE55 pKa = 4.55SGAVAACPGGWACAGVRR72 pKa = 11.84LVVDD76 pKa = 4.12PASVQVRR83 pKa = 11.84AVSTDD88 pKa = 2.93VSLDD92 pKa = 3.04GSLRR96 pKa = 11.84TLRR99 pKa = 11.84CKK101 pKa = 10.55ARR103 pKa = 11.84AIVAIAPSDD112 pKa = 3.38QTDD115 pKa = 3.83RR116 pKa = 11.84IGPIFDD122 pKa = 4.0AAVQVAQRR130 pKa = 11.84EE131 pKa = 4.33LGVLGVVGLARR142 pKa = 11.84LAVSWHH148 pKa = 5.16GMAEE152 pKa = 4.02QVRR155 pKa = 11.84VLKK158 pKa = 10.53PGEE161 pKa = 4.2GSEE164 pKa = 4.55KK165 pKa = 10.37DD166 pKa = 3.31TT167 pKa = 4.86
MM1 pKa = 7.34SGKK4 pKa = 10.33SFVIQTEE11 pKa = 4.31CEE13 pKa = 4.19GVGRR17 pKa = 11.84RR18 pKa = 11.84GLVSASVMEE27 pKa = 4.68FYY29 pKa = 10.84FIIGVVEE36 pKa = 4.41DD37 pKa = 3.71VCQEE41 pKa = 3.87VATRR45 pKa = 11.84LACLEE50 pKa = 4.31AEE52 pKa = 4.59FKK54 pKa = 11.01EE55 pKa = 4.55SGAVAACPGGWACAGVRR72 pKa = 11.84LVVDD76 pKa = 4.12PASVQVRR83 pKa = 11.84AVSTDD88 pKa = 2.93VSLDD92 pKa = 3.04GSLRR96 pKa = 11.84TLRR99 pKa = 11.84CKK101 pKa = 10.55ARR103 pKa = 11.84AIVAIAPSDD112 pKa = 3.38QTDD115 pKa = 3.83RR116 pKa = 11.84IGPIFDD122 pKa = 4.0AAVQVAQRR130 pKa = 11.84EE131 pKa = 4.33LGVLGVVGLARR142 pKa = 11.84LAVSWHH148 pKa = 5.16GMAEE152 pKa = 4.02QVRR155 pKa = 11.84VLKK158 pKa = 10.53PGEE161 pKa = 4.2GSEE164 pKa = 4.55KK165 pKa = 10.37DD166 pKa = 3.31TT167 pKa = 4.86
Molecular weight: 17.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A513ZVE6|A0A513ZVE6_9VIRU Putative capsid protein OS=Fusarium redolens polymycovirus 1 OX=2546034 PE=4 SV=1
MM1 pKa = 7.8ASTTSSVQEE10 pKa = 3.77WAFSVVDD17 pKa = 3.37EE18 pKa = 4.4RR19 pKa = 11.84TRR21 pKa = 11.84VIPEE25 pKa = 3.69RR26 pKa = 11.84HH27 pKa = 4.94VVLPHH32 pKa = 5.85GEE34 pKa = 4.07RR35 pKa = 11.84AILPTPAPGSSVTTAQHH52 pKa = 5.57RR53 pKa = 11.84TPSEE57 pKa = 3.29IRR59 pKa = 11.84ADD61 pKa = 3.49HH62 pKa = 6.74AGLTEE67 pKa = 3.77AVRR70 pKa = 11.84RR71 pKa = 11.84VSLQGEE77 pKa = 4.44SRR79 pKa = 11.84RR80 pKa = 11.84TTDD83 pKa = 2.57VGLRR87 pKa = 11.84GAAHH91 pKa = 6.0SRR93 pKa = 11.84VSRR96 pKa = 11.84ASVRR100 pKa = 11.84PAATEE105 pKa = 3.88EE106 pKa = 4.28GASAVSSRR114 pKa = 11.84RR115 pKa = 11.84SEE117 pKa = 4.03NVGRR121 pKa = 11.84TGEE124 pKa = 4.35SVTTLPVYY132 pKa = 10.56RR133 pKa = 11.84SAEE136 pKa = 3.98EE137 pKa = 3.97LASSNIPQRR146 pKa = 11.84GDD148 pKa = 3.76ANVLWRR154 pKa = 11.84DD155 pKa = 3.26QTVRR159 pKa = 11.84GDD161 pKa = 3.77LEE163 pKa = 4.77SPVHH167 pKa = 6.42SGATGIPRR175 pKa = 11.84VASTATNQQEE185 pKa = 4.25GSYY188 pKa = 10.85GPEE191 pKa = 3.62NVLDD195 pKa = 4.04FQVPPTYY202 pKa = 10.79VGVPGAAVGNTGGGGSEE219 pKa = 3.73PTVVVEE225 pKa = 4.54RR226 pKa = 11.84PRR228 pKa = 11.84GRR230 pKa = 11.84RR231 pKa = 11.84SVRR234 pKa = 11.84GTRR237 pKa = 11.84RR238 pKa = 11.84ATDD241 pKa = 3.35SVGVLTSALAALPEE255 pKa = 4.71GSSFTISVAEE265 pKa = 4.05EE266 pKa = 3.78VAPDD270 pKa = 3.69GTARR274 pKa = 11.84SQASVSVTASSRR286 pKa = 11.84ASGSTSSRR294 pKa = 11.84RR295 pKa = 11.84VRR297 pKa = 11.84EE298 pKa = 3.85VGDD301 pKa = 3.39GHH303 pKa = 5.6EE304 pKa = 4.13HH305 pKa = 5.08HH306 pKa = 6.9HH307 pKa = 6.35RR308 pKa = 11.84RR309 pKa = 11.84HH310 pKa = 5.28KK311 pKa = 10.26RR312 pKa = 11.84DD313 pKa = 3.34RR314 pKa = 11.84RR315 pKa = 11.84DD316 pKa = 3.22SGVTFAEE323 pKa = 4.5WVAGRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 10.04
MM1 pKa = 7.8ASTTSSVQEE10 pKa = 3.77WAFSVVDD17 pKa = 3.37EE18 pKa = 4.4RR19 pKa = 11.84TRR21 pKa = 11.84VIPEE25 pKa = 3.69RR26 pKa = 11.84HH27 pKa = 4.94VVLPHH32 pKa = 5.85GEE34 pKa = 4.07RR35 pKa = 11.84AILPTPAPGSSVTTAQHH52 pKa = 5.57RR53 pKa = 11.84TPSEE57 pKa = 3.29IRR59 pKa = 11.84ADD61 pKa = 3.49HH62 pKa = 6.74AGLTEE67 pKa = 3.77AVRR70 pKa = 11.84RR71 pKa = 11.84VSLQGEE77 pKa = 4.44SRR79 pKa = 11.84RR80 pKa = 11.84TTDD83 pKa = 2.57VGLRR87 pKa = 11.84GAAHH91 pKa = 6.0SRR93 pKa = 11.84VSRR96 pKa = 11.84ASVRR100 pKa = 11.84PAATEE105 pKa = 3.88EE106 pKa = 4.28GASAVSSRR114 pKa = 11.84RR115 pKa = 11.84SEE117 pKa = 4.03NVGRR121 pKa = 11.84TGEE124 pKa = 4.35SVTTLPVYY132 pKa = 10.56RR133 pKa = 11.84SAEE136 pKa = 3.98EE137 pKa = 3.97LASSNIPQRR146 pKa = 11.84GDD148 pKa = 3.76ANVLWRR154 pKa = 11.84DD155 pKa = 3.26QTVRR159 pKa = 11.84GDD161 pKa = 3.77LEE163 pKa = 4.77SPVHH167 pKa = 6.42SGATGIPRR175 pKa = 11.84VASTATNQQEE185 pKa = 4.25GSYY188 pKa = 10.85GPEE191 pKa = 3.62NVLDD195 pKa = 4.04FQVPPTYY202 pKa = 10.79VGVPGAAVGNTGGGGSEE219 pKa = 3.73PTVVVEE225 pKa = 4.54RR226 pKa = 11.84PRR228 pKa = 11.84GRR230 pKa = 11.84RR231 pKa = 11.84SVRR234 pKa = 11.84GTRR237 pKa = 11.84RR238 pKa = 11.84ATDD241 pKa = 3.35SVGVLTSALAALPEE255 pKa = 4.71GSSFTISVAEE265 pKa = 4.05EE266 pKa = 3.78VAPDD270 pKa = 3.69GTARR274 pKa = 11.84SQASVSVTASSRR286 pKa = 11.84ASGSTSSRR294 pKa = 11.84RR295 pKa = 11.84VRR297 pKa = 11.84EE298 pKa = 3.85VGDD301 pKa = 3.39GHH303 pKa = 5.6EE304 pKa = 4.13HH305 pKa = 5.08HH306 pKa = 6.9HH307 pKa = 6.35RR308 pKa = 11.84RR309 pKa = 11.84HH310 pKa = 5.28KK311 pKa = 10.26RR312 pKa = 11.84DD313 pKa = 3.34RR314 pKa = 11.84RR315 pKa = 11.84DD316 pKa = 3.22SGVTFAEE323 pKa = 4.5WVAGRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 10.04
Molecular weight: 34.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3294 |
34 |
771 |
366.0 |
39.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.472 ± 0.853 | 1.791 ± 0.676 |
6.132 ± 0.473 | 5.373 ± 0.427 |
3.491 ± 0.347 | 8.075 ± 0.939 |
2.398 ± 0.318 | 3.279 ± 0.369 |
3.127 ± 0.598 | 7.832 ± 0.864 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.004 ± 0.295 | 2.155 ± 0.246 |
5.556 ± 0.397 | 2.975 ± 0.288 |
8.409 ± 0.75 | 9.624 ± 0.646 |
5.313 ± 0.606 | 9.866 ± 0.765 |
0.88 ± 0.108 | 2.247 ± 0.439 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
