
Candida tropicalis (strain ATCC MYA-3404 / T1) (Yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Candida/Lodderomyces clade; Candida; Candida tropicalis
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
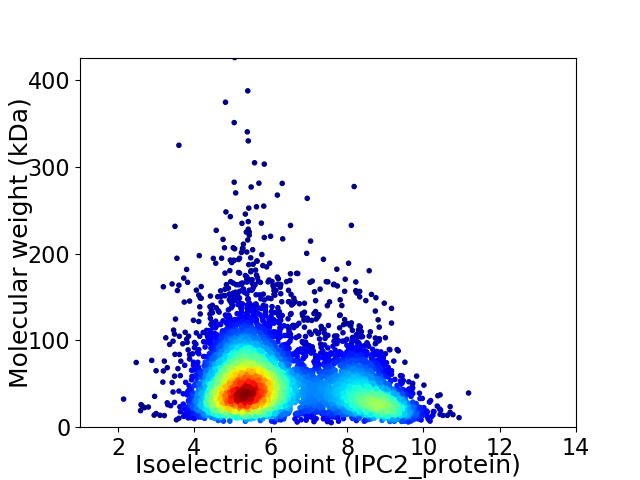
Virtual 2D-PAGE plot for 6226 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
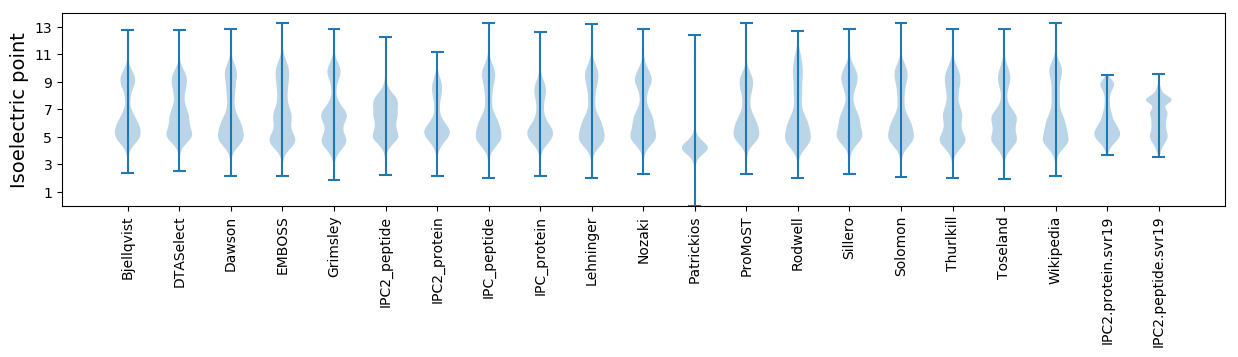
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5M4U6|C5M4U6_CANTT Hyphal_reg_CWP domain-containing protein (Fragment) OS=Candida tropicalis (strain ATCC MYA-3404 / T1) OX=294747 GN=CTRG_01924 PE=4 SV=1
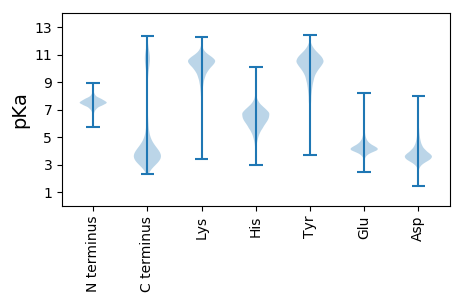
MM1 pKa = 7.57LLLRR5 pKa = 11.84PISLTLVFLFNYY17 pKa = 9.96ALSKK21 pKa = 10.77DD22 pKa = 3.59ITVDD26 pKa = 3.19TVDD29 pKa = 3.92RR30 pKa = 11.84GSISLSVGDD39 pKa = 4.27INIHH43 pKa = 6.13TGASWSIINNAVSALAGSLNVEE65 pKa = 4.21SDD67 pKa = 2.96AGFYY71 pKa = 8.7ITSTSPLIGLDD82 pKa = 3.34VTLLSTLGSIVNDD95 pKa = 4.98GIIAFNSVKK104 pKa = 10.6SLTSSTYY111 pKa = 10.11NLVGAKK117 pKa = 9.94FEE119 pKa = 5.2NNGEE123 pKa = 4.21MYY125 pKa = 10.64LGASGVLSGTMSITSLSWKK144 pKa = 10.43NSGLLVFYY152 pKa = 10.52QNQRR156 pKa = 11.84NSGDD160 pKa = 3.47VRR162 pKa = 11.84LGTPLGSITNDD173 pKa = 3.28GQICLINQVYY183 pKa = 9.59RR184 pKa = 11.84QTNDD188 pKa = 2.54IKK190 pKa = 11.25GSGCITAQKK199 pKa = 10.64NSTIYY204 pKa = 10.21IPSAIQSFDD213 pKa = 3.57TQQSIYY219 pKa = 10.62LKK221 pKa = 10.24DD222 pKa = 3.72SEE224 pKa = 4.82SSIIVQALSTAQTFNVYY241 pKa = 10.52GFGNGNKK248 pKa = 9.48IGLTVPLVGNLIPSYY263 pKa = 9.93PAYY266 pKa = 9.78KK267 pKa = 9.83YY268 pKa = 10.72DD269 pKa = 3.69EE270 pKa = 4.38STGILTLRR278 pKa = 11.84NLLLKK283 pKa = 10.77QNFNIGLGYY292 pKa = 10.22SSSLFQIVTDD302 pKa = 4.06SGIGLPSTLLGSVQYY317 pKa = 10.41NGPVPEE323 pKa = 4.33RR324 pKa = 11.84SISATCQIEE333 pKa = 4.71CKK335 pKa = 9.53EE336 pKa = 4.1LPEE339 pKa = 4.95APGVEE344 pKa = 3.97PTEE347 pKa = 4.07YY348 pKa = 10.1LTTLTSTLSDD358 pKa = 3.25GSVTTEE364 pKa = 3.68SGIVDD369 pKa = 3.75VTTDD373 pKa = 4.2SSGSWYY379 pKa = 6.9TTTSLFPPLSSSSSEE394 pKa = 4.17VVSSSSEE401 pKa = 3.75IVSSTEE407 pKa = 3.98RR408 pKa = 11.84EE409 pKa = 3.87SSLQSFSEE417 pKa = 4.35YY418 pKa = 10.48FNSSSSYY425 pKa = 10.51AFEE428 pKa = 4.2STSSILEE435 pKa = 4.29SSSHH439 pKa = 6.4PSSEE443 pKa = 4.2VSSTQDD449 pKa = 3.37DD450 pKa = 3.69ASEE453 pKa = 5.03SNFSTSSLISEE464 pKa = 4.55SNVLEE469 pKa = 4.39SSDD472 pKa = 3.94ASATSDD478 pKa = 3.3VSSASDD484 pKa = 2.95ISSASDD490 pKa = 3.39ASDD493 pKa = 3.58SDD495 pKa = 3.94PSSTSDD501 pKa = 3.12VSSASDD507 pKa = 2.98ISSASDD513 pKa = 2.88ISSTSEE519 pKa = 3.52IASSSNTSSASDD531 pKa = 2.96ISTSSDD537 pKa = 2.83ASEE540 pKa = 4.63SNASATSGASSASDD554 pKa = 2.94ARR556 pKa = 11.84EE557 pKa = 4.15SNASATSGASDD568 pKa = 3.69ASEE571 pKa = 4.21SDD573 pKa = 3.36ASATSGASDD582 pKa = 3.69ASEE585 pKa = 4.21SDD587 pKa = 3.36ASATSGASDD596 pKa = 3.23ASEE599 pKa = 4.44SNASATSGASDD610 pKa = 3.23ASEE613 pKa = 4.44SNASATSGASSASDD627 pKa = 3.09ASEE630 pKa = 4.01SDD632 pKa = 3.36ASATSGASEE641 pKa = 3.94ASEE644 pKa = 4.53SNASATSGASSASDD658 pKa = 3.04ASSASDD664 pKa = 3.44ANKK667 pKa = 10.61SNAPATSGASDD678 pKa = 3.52ASEE681 pKa = 4.24SDD683 pKa = 3.37ASSTSDD689 pKa = 3.13ASPSYY694 pKa = 10.98NHH696 pKa = 6.91IEE698 pKa = 4.1STEE701 pKa = 4.07SDD703 pKa = 3.58VQAEE707 pKa = 4.43FTTTWTTTNSDD718 pKa = 3.13GSVEE722 pKa = 4.21TEE724 pKa = 3.89SGIVSQSGSSFTTLTTFPPEE744 pKa = 3.78YY745 pKa = 10.12DD746 pKa = 3.59VPSS749 pKa = 3.61
MM1 pKa = 7.57LLLRR5 pKa = 11.84PISLTLVFLFNYY17 pKa = 9.96ALSKK21 pKa = 10.77DD22 pKa = 3.59ITVDD26 pKa = 3.19TVDD29 pKa = 3.92RR30 pKa = 11.84GSISLSVGDD39 pKa = 4.27INIHH43 pKa = 6.13TGASWSIINNAVSALAGSLNVEE65 pKa = 4.21SDD67 pKa = 2.96AGFYY71 pKa = 8.7ITSTSPLIGLDD82 pKa = 3.34VTLLSTLGSIVNDD95 pKa = 4.98GIIAFNSVKK104 pKa = 10.6SLTSSTYY111 pKa = 10.11NLVGAKK117 pKa = 9.94FEE119 pKa = 5.2NNGEE123 pKa = 4.21MYY125 pKa = 10.64LGASGVLSGTMSITSLSWKK144 pKa = 10.43NSGLLVFYY152 pKa = 10.52QNQRR156 pKa = 11.84NSGDD160 pKa = 3.47VRR162 pKa = 11.84LGTPLGSITNDD173 pKa = 3.28GQICLINQVYY183 pKa = 9.59RR184 pKa = 11.84QTNDD188 pKa = 2.54IKK190 pKa = 11.25GSGCITAQKK199 pKa = 10.64NSTIYY204 pKa = 10.21IPSAIQSFDD213 pKa = 3.57TQQSIYY219 pKa = 10.62LKK221 pKa = 10.24DD222 pKa = 3.72SEE224 pKa = 4.82SSIIVQALSTAQTFNVYY241 pKa = 10.52GFGNGNKK248 pKa = 9.48IGLTVPLVGNLIPSYY263 pKa = 9.93PAYY266 pKa = 9.78KK267 pKa = 9.83YY268 pKa = 10.72DD269 pKa = 3.69EE270 pKa = 4.38STGILTLRR278 pKa = 11.84NLLLKK283 pKa = 10.77QNFNIGLGYY292 pKa = 10.22SSSLFQIVTDD302 pKa = 4.06SGIGLPSTLLGSVQYY317 pKa = 10.41NGPVPEE323 pKa = 4.33RR324 pKa = 11.84SISATCQIEE333 pKa = 4.71CKK335 pKa = 9.53EE336 pKa = 4.1LPEE339 pKa = 4.95APGVEE344 pKa = 3.97PTEE347 pKa = 4.07YY348 pKa = 10.1LTTLTSTLSDD358 pKa = 3.25GSVTTEE364 pKa = 3.68SGIVDD369 pKa = 3.75VTTDD373 pKa = 4.2SSGSWYY379 pKa = 6.9TTTSLFPPLSSSSSEE394 pKa = 4.17VVSSSSEE401 pKa = 3.75IVSSTEE407 pKa = 3.98RR408 pKa = 11.84EE409 pKa = 3.87SSLQSFSEE417 pKa = 4.35YY418 pKa = 10.48FNSSSSYY425 pKa = 10.51AFEE428 pKa = 4.2STSSILEE435 pKa = 4.29SSSHH439 pKa = 6.4PSSEE443 pKa = 4.2VSSTQDD449 pKa = 3.37DD450 pKa = 3.69ASEE453 pKa = 5.03SNFSTSSLISEE464 pKa = 4.55SNVLEE469 pKa = 4.39SSDD472 pKa = 3.94ASATSDD478 pKa = 3.3VSSASDD484 pKa = 2.95ISSASDD490 pKa = 3.39ASDD493 pKa = 3.58SDD495 pKa = 3.94PSSTSDD501 pKa = 3.12VSSASDD507 pKa = 2.98ISSASDD513 pKa = 2.88ISSTSEE519 pKa = 3.52IASSSNTSSASDD531 pKa = 2.96ISTSSDD537 pKa = 2.83ASEE540 pKa = 4.63SNASATSGASSASDD554 pKa = 2.94ARR556 pKa = 11.84EE557 pKa = 4.15SNASATSGASDD568 pKa = 3.69ASEE571 pKa = 4.21SDD573 pKa = 3.36ASATSGASDD582 pKa = 3.69ASEE585 pKa = 4.21SDD587 pKa = 3.36ASATSGASDD596 pKa = 3.23ASEE599 pKa = 4.44SNASATSGASDD610 pKa = 3.23ASEE613 pKa = 4.44SNASATSGASSASDD627 pKa = 3.09ASEE630 pKa = 4.01SDD632 pKa = 3.36ASATSGASEE641 pKa = 3.94ASEE644 pKa = 4.53SNASATSGASSASDD658 pKa = 3.04ASSASDD664 pKa = 3.44ANKK667 pKa = 10.61SNAPATSGASDD678 pKa = 3.52ASEE681 pKa = 4.24SDD683 pKa = 3.37ASSTSDD689 pKa = 3.13ASPSYY694 pKa = 10.98NHH696 pKa = 6.91IEE698 pKa = 4.1STEE701 pKa = 4.07SDD703 pKa = 3.58VQAEE707 pKa = 4.43FTTTWTTTNSDD718 pKa = 3.13GSVEE722 pKa = 4.21TEE724 pKa = 3.89SGIVSQSGSSFTTLTTFPPEE744 pKa = 3.78YY745 pKa = 10.12DD746 pKa = 3.59VPSS749 pKa = 3.61
Molecular weight: 76.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5M514|C5M514_CANTT Zn(2)-C6 fungal-type domain-containing protein OS=Candida tropicalis (strain ATCC MYA-3404 / T1) OX=294747 GN=CTRG_01992 PE=4 SV=1
MM1 pKa = 7.35IVEE4 pKa = 5.08RR5 pKa = 11.84IKK7 pKa = 10.43MAIQFQVIVLSALIIIISQNWINVVLMIRR36 pKa = 11.84IRR38 pKa = 11.84LSHH41 pKa = 5.57VVIGHH46 pKa = 5.13GTEE49 pKa = 3.78SQYY52 pKa = 11.42LVPHH56 pKa = 5.89QLLPVKK62 pKa = 10.32FHH64 pKa = 5.91HH65 pKa = 6.31HH66 pKa = 4.18QVKK69 pKa = 9.97KK70 pKa = 9.85FHH72 pKa = 6.12HH73 pKa = 6.38HH74 pKa = 4.83PVKK77 pKa = 10.38RR78 pKa = 11.84FHH80 pKa = 6.09LHH82 pKa = 3.58QVKK85 pKa = 9.94RR86 pKa = 11.84FHH88 pKa = 6.64LHH90 pKa = 4.78HH91 pKa = 6.11QRR93 pKa = 11.84KK94 pKa = 7.87FHH96 pKa = 5.28QAPVKK101 pKa = 10.12RR102 pKa = 11.84SHH104 pKa = 5.4QAPVKK109 pKa = 10.07RR110 pKa = 11.84SHH112 pKa = 5.34QVPVKK117 pKa = 10.16RR118 pKa = 11.84FHH120 pKa = 6.01LHH122 pKa = 3.96QVKK125 pKa = 9.94KK126 pKa = 9.51FHH128 pKa = 5.9QAPVKK133 pKa = 10.18KK134 pKa = 10.11FHH136 pKa = 6.22LHH138 pKa = 4.7PVKK141 pKa = 10.39RR142 pKa = 11.84FHH144 pKa = 6.82LHH146 pKa = 4.81HH147 pKa = 6.11QRR149 pKa = 11.84KK150 pKa = 7.87FHH152 pKa = 5.28QAPVKK157 pKa = 10.2RR158 pKa = 11.84SHH160 pKa = 5.34QVPVKK165 pKa = 10.0RR166 pKa = 11.84FHH168 pKa = 5.98HH169 pKa = 6.16HH170 pKa = 4.8PVKK173 pKa = 10.44RR174 pKa = 11.84FHH176 pKa = 6.67LHH178 pKa = 4.81HH179 pKa = 6.11QRR181 pKa = 11.84KK182 pKa = 7.87FHH184 pKa = 5.28QAPVKK189 pKa = 10.2RR190 pKa = 11.84SHH192 pKa = 5.47QVPVKK197 pKa = 10.13KK198 pKa = 10.23FHH200 pKa = 6.04LRR202 pKa = 11.84QVKK205 pKa = 9.87RR206 pKa = 11.84SHH208 pKa = 5.15QAPVKK213 pKa = 10.07RR214 pKa = 11.84SHH216 pKa = 5.34QVPVKK221 pKa = 10.16RR222 pKa = 11.84FHH224 pKa = 5.92LHH226 pKa = 3.58QVKK229 pKa = 10.08RR230 pKa = 11.84SHH232 pKa = 5.39QAPVKK237 pKa = 10.07RR238 pKa = 11.84SHH240 pKa = 5.34QVPVKK245 pKa = 10.16RR246 pKa = 11.84FHH248 pKa = 6.01LHH250 pKa = 3.96QVKK253 pKa = 9.94KK254 pKa = 9.51FHH256 pKa = 5.9QAPVKK261 pKa = 10.18KK262 pKa = 10.11FHH264 pKa = 6.22LHH266 pKa = 4.7PVKK269 pKa = 10.32RR270 pKa = 11.84FHH272 pKa = 6.16LHH274 pKa = 3.58QVKK277 pKa = 9.88RR278 pKa = 11.84FHH280 pKa = 6.04LHH282 pKa = 3.58QVKK285 pKa = 10.05RR286 pKa = 11.84SHH288 pKa = 5.84LHH290 pKa = 3.74QVKK293 pKa = 9.99RR294 pKa = 11.84SHH296 pKa = 5.84LHH298 pKa = 3.71QVKK301 pKa = 9.97RR302 pKa = 11.84SHH304 pKa = 5.39QAPVKK309 pKa = 10.07RR310 pKa = 11.84SHH312 pKa = 5.38QVPVKK317 pKa = 10.26KK318 pKa = 10.84VPTKK322 pKa = 10.44FQQ324 pKa = 3.37
MM1 pKa = 7.35IVEE4 pKa = 5.08RR5 pKa = 11.84IKK7 pKa = 10.43MAIQFQVIVLSALIIIISQNWINVVLMIRR36 pKa = 11.84IRR38 pKa = 11.84LSHH41 pKa = 5.57VVIGHH46 pKa = 5.13GTEE49 pKa = 3.78SQYY52 pKa = 11.42LVPHH56 pKa = 5.89QLLPVKK62 pKa = 10.32FHH64 pKa = 5.91HH65 pKa = 6.31HH66 pKa = 4.18QVKK69 pKa = 9.97KK70 pKa = 9.85FHH72 pKa = 6.12HH73 pKa = 6.38HH74 pKa = 4.83PVKK77 pKa = 10.38RR78 pKa = 11.84FHH80 pKa = 6.09LHH82 pKa = 3.58QVKK85 pKa = 9.94RR86 pKa = 11.84FHH88 pKa = 6.64LHH90 pKa = 4.78HH91 pKa = 6.11QRR93 pKa = 11.84KK94 pKa = 7.87FHH96 pKa = 5.28QAPVKK101 pKa = 10.12RR102 pKa = 11.84SHH104 pKa = 5.4QAPVKK109 pKa = 10.07RR110 pKa = 11.84SHH112 pKa = 5.34QVPVKK117 pKa = 10.16RR118 pKa = 11.84FHH120 pKa = 6.01LHH122 pKa = 3.96QVKK125 pKa = 9.94KK126 pKa = 9.51FHH128 pKa = 5.9QAPVKK133 pKa = 10.18KK134 pKa = 10.11FHH136 pKa = 6.22LHH138 pKa = 4.7PVKK141 pKa = 10.39RR142 pKa = 11.84FHH144 pKa = 6.82LHH146 pKa = 4.81HH147 pKa = 6.11QRR149 pKa = 11.84KK150 pKa = 7.87FHH152 pKa = 5.28QAPVKK157 pKa = 10.2RR158 pKa = 11.84SHH160 pKa = 5.34QVPVKK165 pKa = 10.0RR166 pKa = 11.84FHH168 pKa = 5.98HH169 pKa = 6.16HH170 pKa = 4.8PVKK173 pKa = 10.44RR174 pKa = 11.84FHH176 pKa = 6.67LHH178 pKa = 4.81HH179 pKa = 6.11QRR181 pKa = 11.84KK182 pKa = 7.87FHH184 pKa = 5.28QAPVKK189 pKa = 10.2RR190 pKa = 11.84SHH192 pKa = 5.47QVPVKK197 pKa = 10.13KK198 pKa = 10.23FHH200 pKa = 6.04LRR202 pKa = 11.84QVKK205 pKa = 9.87RR206 pKa = 11.84SHH208 pKa = 5.15QAPVKK213 pKa = 10.07RR214 pKa = 11.84SHH216 pKa = 5.34QVPVKK221 pKa = 10.16RR222 pKa = 11.84FHH224 pKa = 5.92LHH226 pKa = 3.58QVKK229 pKa = 10.08RR230 pKa = 11.84SHH232 pKa = 5.39QAPVKK237 pKa = 10.07RR238 pKa = 11.84SHH240 pKa = 5.34QVPVKK245 pKa = 10.16RR246 pKa = 11.84FHH248 pKa = 6.01LHH250 pKa = 3.96QVKK253 pKa = 9.94KK254 pKa = 9.51FHH256 pKa = 5.9QAPVKK261 pKa = 10.18KK262 pKa = 10.11FHH264 pKa = 6.22LHH266 pKa = 4.7PVKK269 pKa = 10.32RR270 pKa = 11.84FHH272 pKa = 6.16LHH274 pKa = 3.58QVKK277 pKa = 9.88RR278 pKa = 11.84FHH280 pKa = 6.04LHH282 pKa = 3.58QVKK285 pKa = 10.05RR286 pKa = 11.84SHH288 pKa = 5.84LHH290 pKa = 3.74QVKK293 pKa = 9.99RR294 pKa = 11.84SHH296 pKa = 5.84LHH298 pKa = 3.71QVKK301 pKa = 9.97RR302 pKa = 11.84SHH304 pKa = 5.39QAPVKK309 pKa = 10.07RR310 pKa = 11.84SHH312 pKa = 5.38QVPVKK317 pKa = 10.26KK318 pKa = 10.84VPTKK322 pKa = 10.44FQQ324 pKa = 3.37
Molecular weight: 39.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3018650 |
50 |
3808 |
484.8 |
54.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.05 ± 0.037 | 1.101 ± 0.009 |
5.983 ± 0.026 | 6.759 ± 0.033 |
4.491 ± 0.021 | 5.091 ± 0.033 |
2.072 ± 0.013 | 7.064 ± 0.026 |
7.222 ± 0.038 | 9.14 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.882 ± 0.011 | 6.269 ± 0.027 |
4.553 ± 0.03 | 4.272 ± 0.025 |
3.821 ± 0.018 | 9.06 ± 0.045 |
5.992 ± 0.039 | 5.623 ± 0.024 |
0.984 ± 0.01 | 3.571 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
