
Palaemonetes kadiakensis Mississippi grass shrimp associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 9.08
Get precalculated fractions of proteins
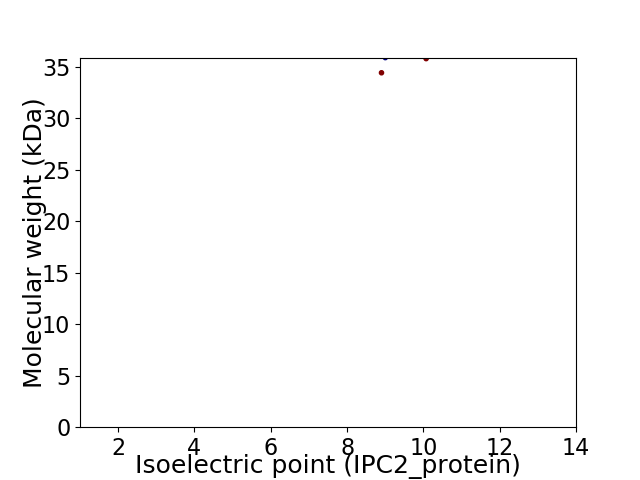
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
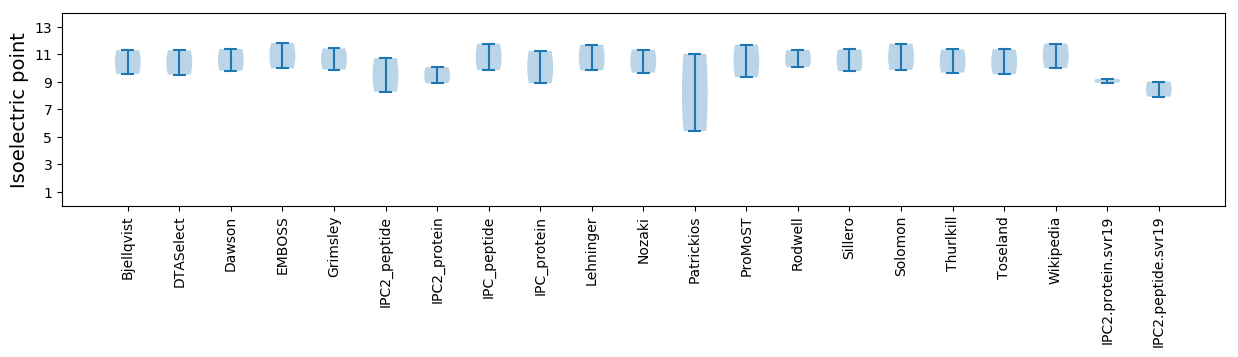
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL61|A0A0K1RL61_9CIRC Putative capsid protein OS=Palaemonetes kadiakensis Mississippi grass shrimp associated circular virus OX=1692258 PE=4 SV=1
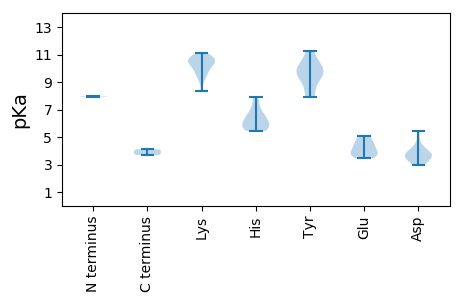
MM1 pKa = 7.98RR2 pKa = 11.84EE3 pKa = 3.8KK4 pKa = 10.72EE5 pKa = 4.35RR6 pKa = 11.84NNTAAPHH13 pKa = 5.97EE14 pKa = 4.55PVSRR18 pKa = 11.84DD19 pKa = 3.15PGSIRR24 pKa = 11.84RR25 pKa = 11.84TKK27 pKa = 10.24SIKK30 pKa = 9.74RR31 pKa = 11.84DD32 pKa = 3.39AVSLSRR38 pKa = 11.84RR39 pKa = 11.84VARR42 pKa = 11.84WWLLTIPEE50 pKa = 4.99DD51 pKa = 4.14DD52 pKa = 3.9WNPPADD58 pKa = 3.88LPAGVAHH65 pKa = 7.39LKK67 pKa = 10.39GQLEE71 pKa = 4.42SGAEE75 pKa = 3.47GGYY78 pKa = 9.22RR79 pKa = 11.84HH80 pKa = 5.73WQVMVSFTTQVRR92 pKa = 11.84LPAVKK97 pKa = 10.34ALFSRR102 pKa = 11.84TAHH105 pKa = 6.41CEE107 pKa = 3.64PSRR110 pKa = 11.84SDD112 pKa = 4.4AVDD115 pKa = 3.28KK116 pKa = 10.42YY117 pKa = 9.94VWKK120 pKa = 10.36EE121 pKa = 3.74DD122 pKa = 3.21TRR124 pKa = 11.84VPGTQFEE131 pKa = 4.61LGRR134 pKa = 11.84APVKK138 pKa = 10.76LNSKK142 pKa = 9.45PDD144 pKa = 3.08WDD146 pKa = 5.42AIWDD150 pKa = 3.68AAKK153 pKa = 10.52AGNIEE158 pKa = 5.09AIPAGIRR165 pKa = 11.84VRR167 pKa = 11.84SYY169 pKa = 11.27ANLRR173 pKa = 11.84RR174 pKa = 11.84IRR176 pKa = 11.84SDD178 pKa = 3.06HH179 pKa = 6.49AVALPVEE186 pKa = 4.77RR187 pKa = 11.84SCRR190 pKa = 11.84VFVGPTGTGKK200 pKa = 8.31SHH202 pKa = 6.92RR203 pKa = 11.84AWSEE207 pKa = 3.55AGMDD211 pKa = 4.88AYY213 pKa = 10.12PKK215 pKa = 10.5DD216 pKa = 3.81PRR218 pKa = 11.84TKK220 pKa = 9.95FWDD223 pKa = 4.42GYY225 pKa = 10.21RR226 pKa = 11.84DD227 pKa = 3.63QLHH230 pKa = 5.45VVIDD234 pKa = 3.87EE235 pKa = 4.08FRR237 pKa = 11.84GGIDD241 pKa = 2.99IAHH244 pKa = 7.88LLRR247 pKa = 11.84WLDD250 pKa = 3.59RR251 pKa = 11.84YY252 pKa = 9.34PVNVEE257 pKa = 3.52IKK259 pKa = 9.72GASMPLAAKK268 pKa = 9.22GIWITSNLAPNLWYY282 pKa = 8.39PAPQHH287 pKa = 5.76TATLYY292 pKa = 10.89HH293 pKa = 6.46PGIRR297 pKa = 11.84YY298 pKa = 7.98WSKK301 pKa = 10.82CCAEE305 pKa = 4.11
MM1 pKa = 7.98RR2 pKa = 11.84EE3 pKa = 3.8KK4 pKa = 10.72EE5 pKa = 4.35RR6 pKa = 11.84NNTAAPHH13 pKa = 5.97EE14 pKa = 4.55PVSRR18 pKa = 11.84DD19 pKa = 3.15PGSIRR24 pKa = 11.84RR25 pKa = 11.84TKK27 pKa = 10.24SIKK30 pKa = 9.74RR31 pKa = 11.84DD32 pKa = 3.39AVSLSRR38 pKa = 11.84RR39 pKa = 11.84VARR42 pKa = 11.84WWLLTIPEE50 pKa = 4.99DD51 pKa = 4.14DD52 pKa = 3.9WNPPADD58 pKa = 3.88LPAGVAHH65 pKa = 7.39LKK67 pKa = 10.39GQLEE71 pKa = 4.42SGAEE75 pKa = 3.47GGYY78 pKa = 9.22RR79 pKa = 11.84HH80 pKa = 5.73WQVMVSFTTQVRR92 pKa = 11.84LPAVKK97 pKa = 10.34ALFSRR102 pKa = 11.84TAHH105 pKa = 6.41CEE107 pKa = 3.64PSRR110 pKa = 11.84SDD112 pKa = 4.4AVDD115 pKa = 3.28KK116 pKa = 10.42YY117 pKa = 9.94VWKK120 pKa = 10.36EE121 pKa = 3.74DD122 pKa = 3.21TRR124 pKa = 11.84VPGTQFEE131 pKa = 4.61LGRR134 pKa = 11.84APVKK138 pKa = 10.76LNSKK142 pKa = 9.45PDD144 pKa = 3.08WDD146 pKa = 5.42AIWDD150 pKa = 3.68AAKK153 pKa = 10.52AGNIEE158 pKa = 5.09AIPAGIRR165 pKa = 11.84VRR167 pKa = 11.84SYY169 pKa = 11.27ANLRR173 pKa = 11.84RR174 pKa = 11.84IRR176 pKa = 11.84SDD178 pKa = 3.06HH179 pKa = 6.49AVALPVEE186 pKa = 4.77RR187 pKa = 11.84SCRR190 pKa = 11.84VFVGPTGTGKK200 pKa = 8.31SHH202 pKa = 6.92RR203 pKa = 11.84AWSEE207 pKa = 3.55AGMDD211 pKa = 4.88AYY213 pKa = 10.12PKK215 pKa = 10.5DD216 pKa = 3.81PRR218 pKa = 11.84TKK220 pKa = 9.95FWDD223 pKa = 4.42GYY225 pKa = 10.21RR226 pKa = 11.84DD227 pKa = 3.63QLHH230 pKa = 5.45VVIDD234 pKa = 3.87EE235 pKa = 4.08FRR237 pKa = 11.84GGIDD241 pKa = 2.99IAHH244 pKa = 7.88LLRR247 pKa = 11.84WLDD250 pKa = 3.59RR251 pKa = 11.84YY252 pKa = 9.34PVNVEE257 pKa = 3.52IKK259 pKa = 9.72GASMPLAAKK268 pKa = 9.22GIWITSNLAPNLWYY282 pKa = 8.39PAPQHH287 pKa = 5.76TATLYY292 pKa = 10.89HH293 pKa = 6.46PGIRR297 pKa = 11.84YY298 pKa = 7.98WSKK301 pKa = 10.82CCAEE305 pKa = 4.11
Molecular weight: 34.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL61|A0A0K1RL61_9CIRC Putative capsid protein OS=Palaemonetes kadiakensis Mississippi grass shrimp associated circular virus OX=1692258 PE=4 SV=1
MM1 pKa = 7.94PGTLVRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84SVGWARR17 pKa = 11.84RR18 pKa = 11.84AGAAVSVARR27 pKa = 11.84GVWRR31 pKa = 11.84GVKK34 pKa = 9.54RR35 pKa = 11.84VRR37 pKa = 11.84SAISRR42 pKa = 11.84GRR44 pKa = 11.84SLARR48 pKa = 11.84STSRR52 pKa = 11.84TRR54 pKa = 11.84SKK56 pKa = 10.83SRR58 pKa = 11.84PKK60 pKa = 10.63SSTRR64 pKa = 11.84PSSAAPMSVDD74 pKa = 3.53PQSISHH80 pKa = 6.48GSVSLRR86 pKa = 11.84LASPVKK92 pKa = 10.4GMKK95 pKa = 10.13SLGRR99 pKa = 11.84WVYY102 pKa = 7.9TQQNNGRR109 pKa = 11.84IVAGLGLQGAGVLLGHH125 pKa = 5.86NTISQLVVDD134 pKa = 4.18TAVPNGTQFKK144 pKa = 10.86DD145 pKa = 3.7DD146 pKa = 3.79VFAMNPYY153 pKa = 9.09QANTGSGIFGSILAPSQDD171 pKa = 3.49RR172 pKa = 11.84IMCRR176 pKa = 11.84SVRR179 pKa = 11.84TKK181 pKa = 10.86FMMANTSNAPCVVVLYY197 pKa = 9.7WVLSKK202 pKa = 10.57KK203 pKa = 10.11AHH205 pKa = 5.84QGNPFDD211 pKa = 3.7RR212 pKa = 11.84WTSFLSEE219 pKa = 4.08TALGQSTATQPNQSDD234 pKa = 3.87SSTGPGTTGVPTFAVYY250 pKa = 9.76GQEE253 pKa = 4.0PTTVRR258 pKa = 11.84SWRR261 pKa = 11.84NTFKK265 pKa = 11.12LLRR268 pKa = 11.84RR269 pKa = 11.84KK270 pKa = 9.15EE271 pKa = 3.92YY272 pKa = 10.48HH273 pKa = 5.7MSFGAVQCINYY284 pKa = 9.55SVAVNKK290 pKa = 9.86MFDD293 pKa = 3.38KK294 pKa = 10.91TVATQHH300 pKa = 5.47NTLTSTGYY308 pKa = 9.86PGGTVWLCAVVRR320 pKa = 11.84GTRR323 pKa = 11.84DD324 pKa = 3.44LVPDD328 pKa = 4.07CWW330 pKa = 3.7
MM1 pKa = 7.94PGTLVRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84SVGWARR17 pKa = 11.84RR18 pKa = 11.84AGAAVSVARR27 pKa = 11.84GVWRR31 pKa = 11.84GVKK34 pKa = 9.54RR35 pKa = 11.84VRR37 pKa = 11.84SAISRR42 pKa = 11.84GRR44 pKa = 11.84SLARR48 pKa = 11.84STSRR52 pKa = 11.84TRR54 pKa = 11.84SKK56 pKa = 10.83SRR58 pKa = 11.84PKK60 pKa = 10.63SSTRR64 pKa = 11.84PSSAAPMSVDD74 pKa = 3.53PQSISHH80 pKa = 6.48GSVSLRR86 pKa = 11.84LASPVKK92 pKa = 10.4GMKK95 pKa = 10.13SLGRR99 pKa = 11.84WVYY102 pKa = 7.9TQQNNGRR109 pKa = 11.84IVAGLGLQGAGVLLGHH125 pKa = 5.86NTISQLVVDD134 pKa = 4.18TAVPNGTQFKK144 pKa = 10.86DD145 pKa = 3.7DD146 pKa = 3.79VFAMNPYY153 pKa = 9.09QANTGSGIFGSILAPSQDD171 pKa = 3.49RR172 pKa = 11.84IMCRR176 pKa = 11.84SVRR179 pKa = 11.84TKK181 pKa = 10.86FMMANTSNAPCVVVLYY197 pKa = 9.7WVLSKK202 pKa = 10.57KK203 pKa = 10.11AHH205 pKa = 5.84QGNPFDD211 pKa = 3.7RR212 pKa = 11.84WTSFLSEE219 pKa = 4.08TALGQSTATQPNQSDD234 pKa = 3.87SSTGPGTTGVPTFAVYY250 pKa = 9.76GQEE253 pKa = 4.0PTTVRR258 pKa = 11.84SWRR261 pKa = 11.84NTFKK265 pKa = 11.12LLRR268 pKa = 11.84RR269 pKa = 11.84KK270 pKa = 9.15EE271 pKa = 3.92YY272 pKa = 10.48HH273 pKa = 5.7MSFGAVQCINYY284 pKa = 9.55SVAVNKK290 pKa = 9.86MFDD293 pKa = 3.38KK294 pKa = 10.91TVATQHH300 pKa = 5.47NTLTSTGYY308 pKa = 9.86PGGTVWLCAVVRR320 pKa = 11.84GTRR323 pKa = 11.84DD324 pKa = 3.44LVPDD328 pKa = 4.07CWW330 pKa = 3.7
Molecular weight: 35.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
635 |
305 |
330 |
317.5 |
35.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.291 ± 1.111 | 1.417 ± 0.077 |
4.567 ± 1.208 | 2.835 ± 1.514 |
2.52 ± 0.401 | 7.874 ± 0.957 |
2.362 ± 0.666 | 3.622 ± 0.942 |
4.567 ± 0.493 | 6.299 ± 0.188 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.047 ± 0.535 | 3.622 ± 0.488 |
6.299 ± 0.902 | 3.307 ± 0.974 |
9.291 ± 0.158 | 8.976 ± 1.996 |
6.772 ± 1.585 | 8.504 ± 1.176 |
3.307 ± 0.694 | 2.52 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
